Retinitis Pigmentosa 42
A number sign (#) is used with this entry because of evidence that retinitis pigmentosa-42 (RP42) is caused by heterozygous mutation in the KLHL7 gene (611119) on chromosome 7p15.
For a general phenotypic description and a discussion of genetic heterogeneity of retinitis pigmentosa, see 268000.
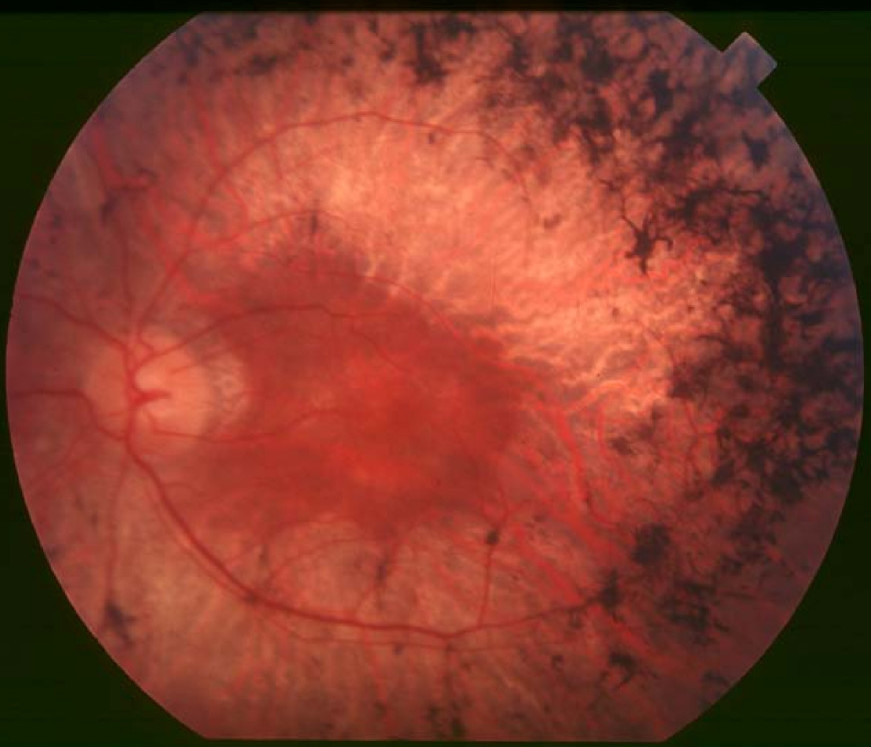
Clinical FeaturesAndreasson (1991) studied 26 affected and 6 unaffected members of 4 families from central Norway and Sweden segregating autosomal dominant retinitis pigmentosa. Although fundus examination was similar in patients from all 4 families, showing narrowed vessels and bone spicule pigment, 2 different types of RP could be distinguished by electroretinography (ERG) based on cone b-wave implicit times, which were normal in 1 family ('family 59') but prolonged in the other 3 families. ERG recordings from 10 RP patients in different age groups from a 6-generation family ('family 72') revealed that cone b-wave amplitude decreased with age but implicit time did not, suggesting that amplitude is useful for monitoring the progression of disease whereas implicit time is more suitable for distinguishing different types of disease. Andreasson (1991) found a correlation between age and the log of the amplitude in family 72, with a progression rate of 7.7% per year, corresponding to a half-life of 8.7 years; this was substantially slower than the previously reported average for all RP cases (18%; Berson et al., 1985). Genetic screening failed to detect mutations in the RHO gene (180380).
Wen et al. (2011) performed a comprehensive assessment of patients from 3 families with RP42 (RFS073, RFS038, and RFS061), previously studied by Friedman et al. (2009) and found to have 3 different heterozygous mutations in the KLHL7 gene (611119.0001-611119.0003, respectively). All fundi showed the characteristic signs of RP, with clear loss of retinal tissue in the periphery, bone spicule pigmentation, arteriolar attenuation, and waxy optic pallor. In general, visual acuity reduction did not manifest until 50 years of age, and best-corrected visual acuity was 20/50 or better in at least 1 eye, up to age 65 years. Visual field restriction was the initial motivation for all 3 probands to seek medical attention. Static and kinetic visual fields showed concentric constriction to central 10 degrees to 20 degrees by age 65 years. Two patients exhibited bilateral visual field retention in the far periphery on Goldmann perimetry testing. Both rod and cone full-field electroretinographic amplitudes were substantially lower than normal, with a decline rate of 3% per year in cone 31-Hz flicker response. Rod and cone activation and inactivation variables were abnormal. Spectral-domain optical coherence tomography indicated retention of foveal inner segment-outer segment junction through age 65 years. Wen et al. (2011) concluded that mutations in KLHL7 are associated with a late-onset form of autosomal dominant retinal degeneration that preferentially affects the rod photoreceptors, and suggested that strong retention of foveal function and bilateral concentric constriction of visual fields with far periphery sparing might guide mutation screening in autosomal dominant RP.
MappingFriedman et al. (2009) performed a whole-genome scan of 23 members of a large 6-generation Scandinavian family with a slow-progressing retinopathy, previously studied by Andreasson (1991) ('family 72'), and obtained a peak multipoint lod score of 5.0 on chromosome 7p15. They designated the locus RP42. Haplotype analysis defined an approximately 3-Mb critical interval between rs4719697 and rs2188993, a region containing 30 annotated or predicted genes. Friedman et al. (2009) noted that the RP42 locus is distinct from the previously reported RP9 locus on chromosome 7p14.2 (180104).
Molecular GeneticsIn a large 6-generation Scandinavian family with a slow-progressing retinopathy mapping to chromosome 7p15, previously studied by Andreasson (1991) (family 72), Friedman et al. (2009) screened several candidate genes and identified a heterozygous mutation in the KLHL7 gene (S150N; 611119.0001) that segregated completely with disease in the 24 family members who were examined. The mutation was not found in 470 controls, including 183 from Scandinavia. Friedman et al. (2009) then screened the KLHL7 gene in 504 unrelated probands with autosomal dominant RP and identified the S150N mutation in a North American family; 2 additional heterozygous missense mutations (611119.0002 and 611119.0003) were identified in 3 families and 1 proband, respectively.