Retinitis Pigmentosa 76
A number sign (#) is used with this entry because of evidence that retinitis pigmentosa-76 (RP76) is caused by homozygous mutation in the POMGNT1 gene (606822) on chromosome 1p34.
For a general phenotypic description and a discussion of genetic heterogeneity of retinitis pigmentosa, see 268000.
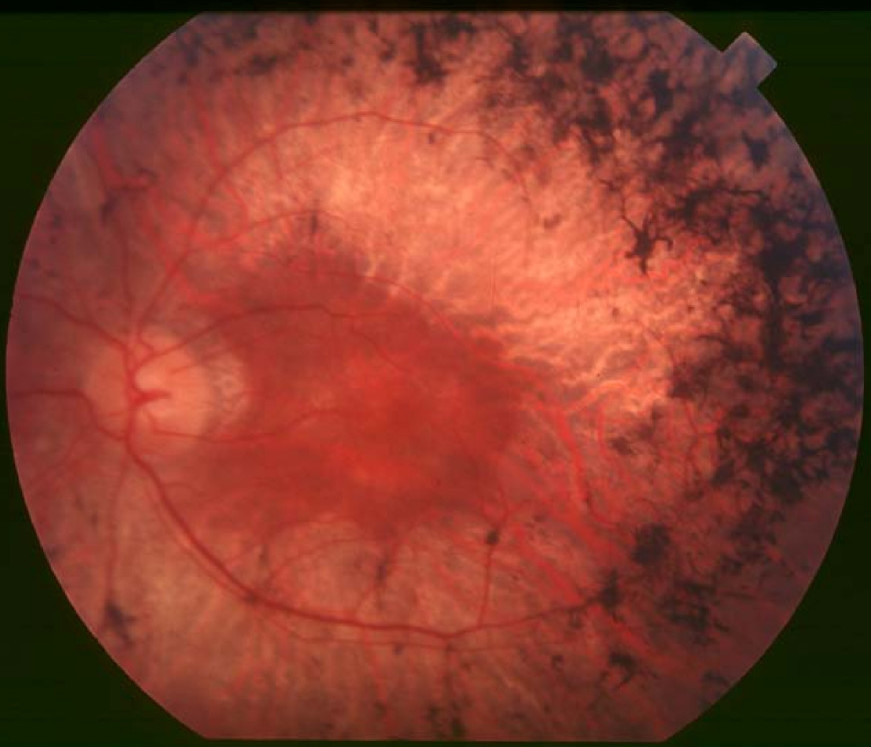
Clinical FeaturesXu et al. (2016) studied an Italian sister and brother with retinitis pigmentosa. The 78-year-old sister, who became aware of nyctalopia and vision problems at age 12 years, showed visual acuity of 20/80 in the right eye and light-perception only in the left eye, constricted visual fields, and peripapillary atrophy; funduscopy revealed bone spicule pigmentation and attenuated retinal vessels. Her 69-year-old brother, who was aware of vision problems at age 10 years, had visual acuities of 20/100 and very restricted visual fields bilaterally, with bone spicules, narrow retinal vessels, peripapillary atrophy, and a tigroid appearance of the fundus. The authors also examined 2 unrelated Han Chinese patients with nonsyndromic RP: a 32-year-old woman who had night blindness since childhood and showed visual acuities of 20/25 and 20/50, with a tigroid appearance of the fundus and constricted visual fields; and a 52-year-old unrelated man who had night blindness since his youth, with visual acuity of 20/400 in the right eye and 20/40 in the left eye, tunnel vision bilaterally, and bone spicule as well as salt and pepper pigmentation scattered throughout the retina, with macular involvement in both eyes. Optical coherence tomography was performed in 3 of the patients and showed retinal thinning in all 3, with absent inner/outer segment junctions in 2; other features included chorioretinal atrophy, flat fovea, and cystoid macular edema. Electroretinography (ERG) in the Han Chinese woman revealed nondetectable waves bilaterally, and fundus autofluorescence showed oval-shaped hypofluorescence with a hyperfluorescent ring in the macula and mottled hypofluorescence in the peripheral retina. None of the 4 patients exhibited any extraocular symptoms.
Wang et al. (2016) studied 17 patients with RP from 5 families belonging to an ethnic minority group living in a closed community on a small island off the southeastern coast of Taiwan. All 17 patients had night blindness, with self-reported age of onset ranging from 12 to 40 years, and all had peripheral visual field loss. Examination showed typical attenuated retinal arterioles, bone spicule pigment deposits, and optic disc pallor; in addition, rod responses were severely reduced on ERG. The patients exhibited no extraocular features; specifically, no mental retardation and no muscle weakness or atrophy was observed.
MappingIn 17 affected individuals from 5 families with nonsyndromic RP from a closed community on a small Taiwanese island, Wang et al. (2016) identified an approximately 1.8-Mb shared homozygous region at chromosome 1p34-p33, bounded by SNPs rs2983715 and rs324423. The authors obtained a maximum lod score of 4.4 for the region and noted that it contained no known RP genes and did not overlap with any previously reported RP loci.
Molecular GeneticsIn a 78-year-old Italian woman with nonsyndromic retinitis pigmentosa, Xu et al. (2016) performed targeted capture sequencing and excluded mutations in known RP-associated genes. Whole-exome sequencing in the woman and her affected brother revealed compound heterozygosity for a missense (I287S; 606822.0018) and a nonsense (R63X; 606822.0019) mutation in the POMGNT1 gene. Screening of exome data from 308 unsolved RP patients for mutations in POMGNT1 identified a 32-year-old Han Chinese woman who was homozygous for a missense mutation (E156K; 606822.0020) and an unrelated 52-year-old Han Chinese man who was compound heterozygous for a truncating (V633X; 606822.0021) and a missense (G502A; 606822.0022) mutation. Homozygosity mapping in the remaining unsolved RP patients revealed 5 probands with homozygous regions that included the POMGNT1 gene; however, Sanger analysis of the entire coding sequence did not reveal any pathogenic mutations in the patients.