Retinitis Pigmentosa 18
A number sign (#) is used with this entry due to evidence that this form of retinitis pigmentosa, designated RP18, is caused by heterozygous mutation in the PRPF3 gene (607301) on chromosome 1q21.
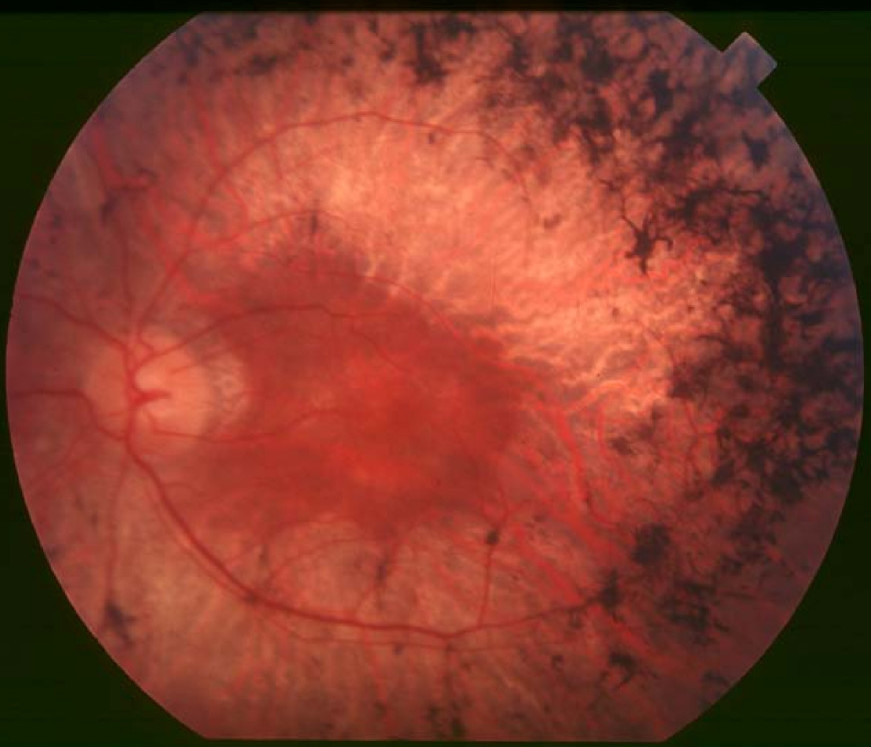
For a phenotypic description and a discussion of genetic heterogeneity of retinitis pigmentosa, see 268000.
Clinical FeaturesXu et al. (1996) studied a large Danish family of 7 generations in which autosomal dominant retinitis pigmentosa segregated. Clinical diagnosis was based on a history of night blindness and ocular fundus findings typical of retinitis pigmentosa and included peripheral bone spicule formation, severe constriction of retinal arterioles, and progressive visual field defects beginning as midperipheral ring scotomas. Pathologic dark adaptation did not occur until the end of the first decade.
MappingIn a 7-generation Danish family with autosomal dominant retinitis pigmentosa, Xu et al. (1996) found linkage without recombination between RP18 and D1S498, which maps to chromosome 1q near the centromere. Analysis of multiple informative meioses suggests that in this family D1S534 and D1S305 flank RP18 in interval 1p13-q23. Xu et al. (1998) refined the genetic mapping of RP18 to a 2-cM region between D1S442 and D1S2858 on 1q.
Inglehearn et al. (1998) reported an English family with autosomal dominant retinitis pigmentosa mapping to this locus. Haplotype analysis placed the locus proximal to D1S1664. This marker is proximal to D1S2346, which is genetically indistinguishable from D1S2858; furthermore, 3 CEPH YACs containing D1S1664 did not contain either D1S2858 or D1S2346. Inglehearn et al. (1998) therefore concluded that their analysis further refined the distal boundary of the RP18 interval.
Molecular GeneticsChakarova et al. (2002) screened the PRPF3 gene, which they called HPRP3, in 3 chromosome 1q-linked RP families. Two different missense mutations in 2 English families, a Danish family, and in 3 RP individuals were identified. One of the mutations (T494M; 607301.0001) was seen repeatedly in apparently unlinked families, raising the possibility of a mutation hotspot. Haplotype analysis with PRPF3 SNPs supported multiple origins for the mutation. The altered amino acids, which are highly conserved in all known PRPF3 orthologs, suggested a major function of that domain in the splicing process. Although PRPF3 appears to be ubiquitously expressed, the authors speculated that a retina-specific splicing element may interact with PRPF3 and generate the rod photoreceptor-specific phenotype.