Retinitis Pigmentosa 82 With Or Without Situs Inversus
A number sign (#) is used with this entry because of evidence that retinitis pigmentosa-82 with or without situs inversus (RP82) is caused by homozygous mutation in the ARL2BP gene (615407) on chromosome 16q13.
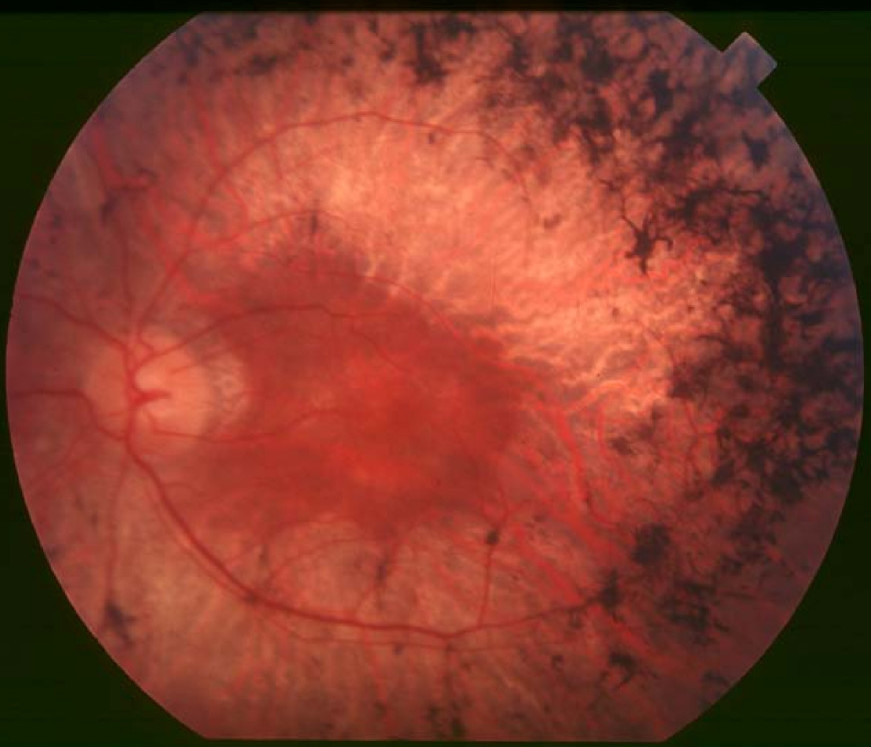
Clinical FeaturesDavidson et al. (2013) studied a consanguineous family of Arab Muslim origin in which 3 sibs were diagnosed with retinitis pigmentosa (RP) in their 20s, after complaints of night vision and visual field impairment. At 36 years of age, the oldest sib could detect hand motion with her right eye and had bare light perception with her left eye, with no significant refractive errors. Mild posterior subcapsular cataracts were present, and funduscopy revealed typical signs of RP, including optic disc pallor, mild to moderate bone-spicule-like pigmentation in the midperiphery, and attenuated retinal blood vessels. All 3 sibs displayed a marked component of macular atrophy, with smaller and larger atrophic patches, as well as epiretinal membranes with wrinkling of the retina. Neither photopic nor scotopic responses were detectable on electroretinography (ERG). Audiometric testing was within normal limits in all 3 sibs. CT imaging revealed full thoracic and abdominal situs inversus in the older sister and brother, whereas their younger brother had normal positioning of the thoracic and abdominal organs.
Audo et al. (2017) reported a consanguineous Moroccan family in which 2 sisters (CIC01154 and CIC01155) were diagnosed in their teens with moderately severe rod-cone dystrophy. At age 36 years, best-corrected visual acuity (BCVA) in 1 sister was 20/200 on the right and counting fingers on the left, with visual fields below 10 degrees. The other sister, at age 27 years, had BCVA of 20/25 in both eyes and visual fields reduced to 30 degrees. Fundus examination showed typical signs of rod-cone dystrophy, including pale optic discs, narrowed retinal vessels, and pigmentary changes in the periphery. There were additional atrophic macular changes in the older sister, and fundus autofluorescence findings were consistent with more advanced disease in the older sister. Chest x-rays did not show situs inversus.
Molecular GeneticsIn 2 sibs with retinitis pigmentosa and full thoracic and abdominal situs inversus from a consanguineous family of Arab Muslim origin, Davidson et al. (2013) performed high-density genomewide SNP microarray analysis followed by whole-exome sequencing, and identified a homozygous splice site mutation in the ARL2BP gene (615407.0001) that segregated with disease in the family and was not present in 100 ethnically matched controls, in the dbSNP or 1000 Genomes Project databases, or in 12,996 control haplotypes from the NHLBI Exome Variant Server. No variants in any known retinal disease-associated genes were identified. In addition, in a 48-year-old man from a consanguineous family of European descent who had been diagnosed with RP and primary ciliary dyskinesia (CILD5; 608647), Davidson et al. (2013) identified homozygosity for a missense mutation in the ARL2BP gene (M45R; 615407.0002) as well as homozygosity for a splice site mutation in the HYDIN gene (610812.0003). No other disease-associated variants were identified in genes previously associated with RP or any other degenerative retinal phenotypes. Bidirectional Sanger sequencing of ARL2BP in 298 unrelated individuals with autosomal recessive retinal degeneration revealed no disease-associated variants, and none were detected in whole-exome sequencing data from 60 additional patients with retinal degeneration.
In 2 sisters from a consanguineous Moroccan family who had severe retinal dystrophy without situs inversus, Audo et al. (2017) identified homozygosity for a splice site mutation in the ARL2BP gene (615407.0003). Their unaffected parents and unaffected sister were heterozygous for the mutation. Analysis of ARL2BP in a total of 844 patients with autosomal recessive rod-cone dystrophy did not reveal any additional pathogenic variants.