Retinitis Pigmentosa 11
A number sign (#) is used with this entry because retinitis pigmentosa-11 (RP11) is caused by heterozygous mutation in the PRPF31 gene (606419) on chromosome 19q13.
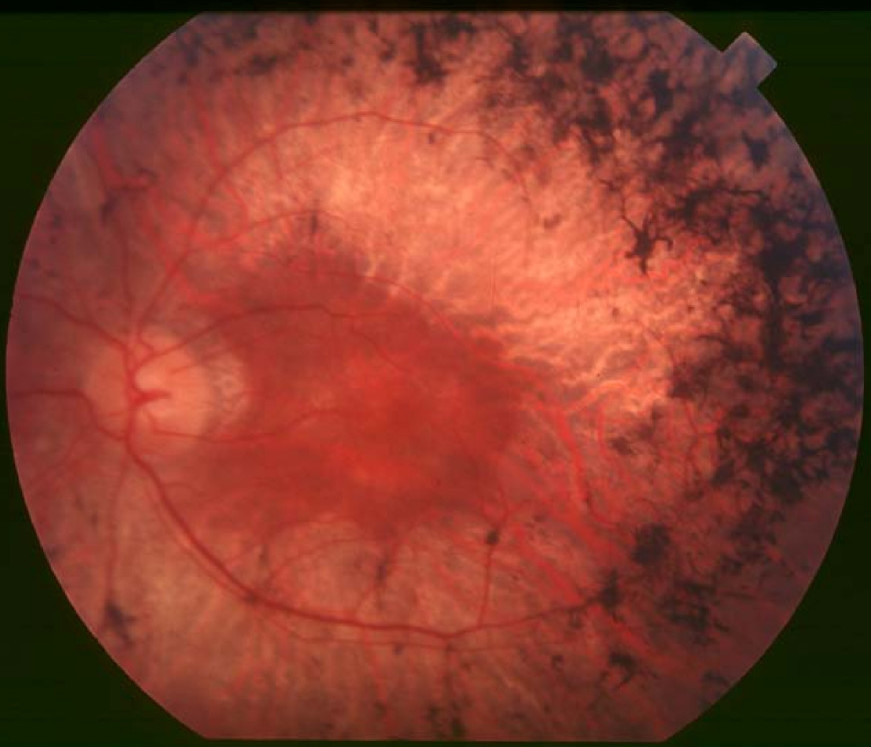
DescriptionRetinitis pigmentosa (RP) is a clinically and genetically heterogeneous group of retinal dystrophies characterized by a progressive degeneration of photoreceptors, eventually resulting in severe visual impairment.
For a discussion of genetic heterogeneity of RP, see 268000.
Clinical FeaturesMoore et al. (1993) described 4 families with autosomal dominant retinitis pigmentosa. Of the 15 patients who were studied from 3 of the families (families 1, 3, and 4), 14 had early onset of night blindness, 10 before age 10 years and 4 before age 20 years, and evidence of severe disease. Patients in all families showed typical fundus features of RP. The majority also had posterior subcapsular lens opacities, macular edema, and/or macular atrophy.
InheritanceThe transmission pattern of retinitis pigmentosa in the families studied by Moore et al. (1993) was consistent with autosomal dominant inheritance.
MappingIn a family (family 4; ADRP5) with retinitis pigmentosa previously reported by Moore et al. (1993), Al-Maghtheh et al. (1994) found linkage of the disorder to chromosome 19q13.4. Three-point analysis of the RP phenotype and markers D19S180 and D19S214 gave a maximum lod score of 4.87. Combining data from these and other markers, Al-Maghtheh et al. (1994) found a lod score of 5.34 in the interval between the 2 markers mentioned, located in the region 19q13.4. Linkage data indicated that this form of retinitis pigmentosa, designated RP11, is separate from the locus for cone-rod dystrophy (120970) mapped to 19q by Evans et al. (1994). Al-Maghtheh et al. (1996) studied families 3 and 4 of Moore et al. (1993) and 2 additional RP families (RP1907 and ADRP2) linked to 19q. They suggested that 19q is a major locus for RP. They were able to refine the RP11 interval to 5 cM between markers D19S180 and AFMc001yb1. All linked families exhibited incomplete penetrance; some obligate gene carriers remained asymptomatic throughout their lives, whereas symptomatic individuals experienced night blindness and visual field loss in their teens and were generally registered as blind by their thirties. Al-Maghtheh et al. (1996) stated that patients with RP mapping to chromosome 19q were either severely affected or asymptomatic, showing an 'all or nothing' form of incomplete penetrance that the authors called 'bimodal expressivity.'
This same entity was apparently identified by Xu et al. (1995) in 4 generations of a Japanese family in which autosomal dominant RP of highly variable expression was segregating. Linkage to D19S180 and other markers on 19q was found. Xu et al. (1995) pointed out that several members of the family reported by Al-Maghtheh et al. (1994) likewise had variable expression; several asymptomatic carriers exhibited functional abnormalities both in electrophysiologic and psychophysical testing.
McGee et al. (1997) studied 3 families with reduced penetrance retinitis pigmentosa, including family W reported by Berson et al. (1969). In all 3 families, the disease gene appeared to be linked to 19q13.4, the region containing the RP11 locus, as defined by previously reported linkage studies based on 5 other reduced penetrance families. Meiotic recombinants in 1 of the newly identified RP11 families and in 2 of the previously reported families served to restrict the disease locus to a 6-cM region bounded by markers D19S574 and D19S926. McGee et al. (1997) also compared the disease status of RP11 carriers with the segregation of microsatellite alleles within 19q13.4 from the noncarrier parents in the newly reported and the previously reported families. The results supported the hypothesis that wildtype alleles at the RP11 locus or at a closely linked locus inherited from noncarrier parents are a major factor influencing the penetrance of pathogenic alleles at this locus. 'Isoallele' is the designation used for an allele that modifies the expression of the disease allele in trans. Another example is provided by the variable severity of elliptocytosis due to mutation in alpha-spectrin of the erythrocyte (182860) depending on the state of the 'normal allele' (Gratzer, 1994). Modification by an isoallele has also been suggested as the explanation for variability of phenotype and unusual inheritance in erythropoietic protoporphyria (177000).
Molecular GeneticsVithana et al. (2001) identified mutations in the PRPF31 gene in families and individuals with RP11. The mutations included missense substitutions, deletions, and insertions (606419.0001-606419.0007).
Wang et al. (2003) described a Chinese kindred with high penetrance of retinitis pigmentosa in association with a 12-bp deletion of PRPF31 (606419.0008). Waseem et al. (2007) noted that RP11 only results from coinheritance of a mutated allele and a wildtype low-expressed allele. They suggested that a high prevalence of low-expressing alleles in certain populations may account for the PRPF31 mutations being identified in patients with RP11 with apparent complete penetrance.
Waseem et al. (2007) identified 6 PRPF31 mutations, 4 of which were novel, in a cohort of 118 patients with autosomal dominant RP in the U.K., including members of family RP1907 described by Al-Maghtheh et al. (1996). The age of onset and the severity of the disease varied with different mutations, and individuals carrying the same mutation showed a range of phenotypic variation, suggesting the involvement of other modifying genes.
In a previously reported family segregating autosomal dominant RP with reduced penetrance (family W in Berson et al., 1969; family 1562 in McGee et al., 1997) in which extensive screening had failed to detect a PRPF31 mutation (McGee et al., 2002; Rivolta et al., 2006), Rio Frio et al. (2009) sequenced the entire PRPF31 genomic region using the Sanger method and ultrahigh-throughput analysis and identified a splice site mutation (606419.0009) that was common to all patients and obligate asymptomatic carriers and was not found in 300 control chromosomes.
HistoryAl-Maghtheh et al. (1998) reported a missense change (arg659 to ser) in the PRKCG gene (176980) in 2 families with RP11-linked dominant RP. Only 1 of the 2 families clearly exhibited the hallmark characteristic of RP11, namely asymptomatic, obligate carriers who transmitted the disease to offspring. Al-Maghtheh et al. (1998) failed to discover a mutation in PRKCG in 3 other families with reduced penetrance showing linkage to this region. Dryja et al. (1999) found no mutations in PRKCG in 3 families that showed linkage to 19q, where both RP11 and PRKCG map, and the characteristic reduced penetrance of RP11. Vithana et al. (2001) stated that the PRKCG gene is not involved in RP11.