Retinitis Pigmentosa 1
A number sign (#) is used with this entry because of evidence that retinitis pigmentosa-1 (RP1) is caused by heterozygous, homozygous, or compound heterozygous mutation in the ORP1 gene (RP1; 603937) on chromosome 8q12.
For a general phenotypic description and a discussion of genetic heterogeneity of retinitis pigmentosa, see 268000.
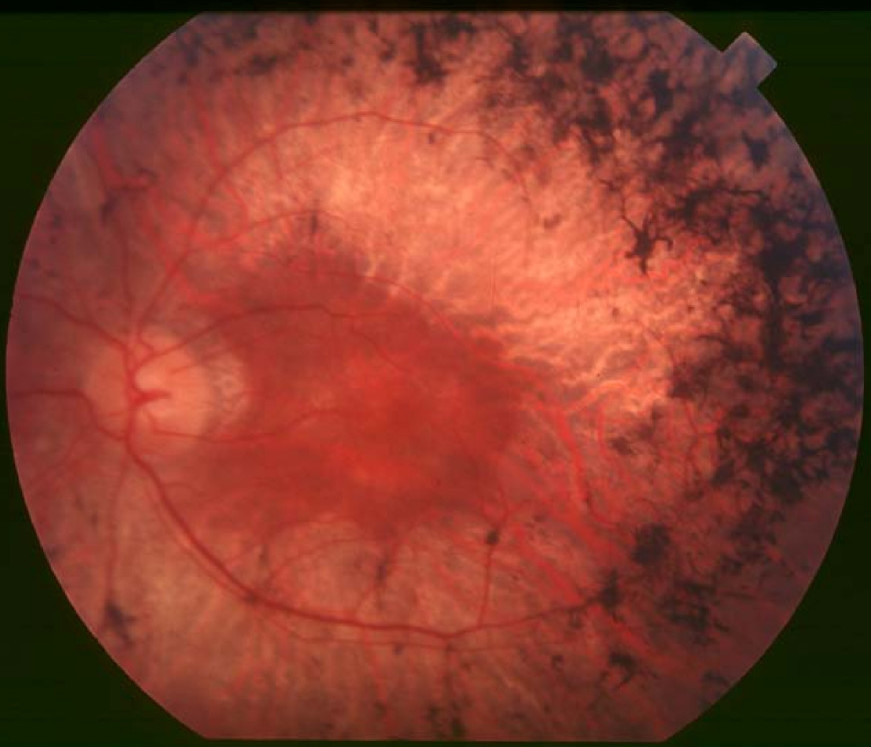
Clinical FeaturesBlanton et al. (1991) described a large extended American family (UCLA-RP01) segregating autosomal dominant retinitis pigmentosa with relatively late onset of night blindness, usually by the third decade of life, and with slow progression. Characteristic clinical findings included diffuse retinal pigmentation, progressive decrease in recordable ERGs, and concentric visual field loss. Funduscopic findings included retinal atrophy, bone-spicule-like pigment deposits, and vascular attenuation. The UCLA-RP01 family had previously been reported by Heckenlively et al. (1982) and Daiger et al. (1989). All of those affected could trace their disease to an affected ancestor living in the early 19th century.
Chassine et al. (2015) determined the refractive error in 26 patients with autosomal recessive RP1 with truncating mutations, 25 with autosomal dominant RP1, 93 with arRP without RP1 mutations, 8 and 33 with X-linked RP2 (312600) and RP3 (300029), respectively, and 198 patients with Usher syndrome (see 276900). Patients with autosomal recessive RP1 and those with X-linked inheritance had moderate to high myopia, whereas patients in the other 3 groups tended to have small refractive errors. The authors suggested that patients with arRP and high myopic refractive error should be preferentially analyzed for RP1 mutations.
MappingBy linkage studies, Blanton et al. (1991) localized the disorder in the ULCA-RP01 family to a locus, designated RP1, in the pericentric region of chromosome 8, approximately 8p11-q21. Exclusion mapping had been useful in narrowing down the search for the locus in the UCLA-RP01 family, as it had been in the mapping of neurofibromatosis (162200), Friedreich ataxia (229300), and Marfan syndrome (154700) loci.
Xu et al. (1996) mapped an adRP locus in an Australian family (family D) to the same region of chromosome 8. Based on the overlap of the linkage data from both families, the critical region for the RP1 locus was limited to approximately 4 cM on 8q11-q13.
Inglehearn et al. (1999) mapped an adRP locus in a family from southwest England to the 8q locus. The phenotype in the British family was similar to those of the American family of Blanton et al. (1991) and the Australian family of Xu et al. (1996). The linkage analysis did not further refine the critical region.
Molecular GeneticsPierce et al. (1999) identified a photoreceptor-specific gene (called RP1, or ORP1 for 'oxygen-regulated photoreceptor protein-1'; 603937) in the same interval on 8q11-q13 to which RP1 had been mapped by Xu et al. (1996). The expression of the gene was modulated by retinal oxygen levels in vivo. In affected members of the UCLA-RP01 family studied by Blanton et al. (1991) and others, Pierce et al. (1999) identified heterozygosity for a nonsense mutation in the RP1 gene (R677X; 603937.0001). Pierce et al. (1999) stated that the R677X mutation was present in approximately 3% of cases of dominant RP in North America. Pierce et al. (1999) also detected 2 deletion mutations that caused frameshifts and introduced premature termination codons in 3 other families with dominant RP (603937.0002 and 603937.0003). Thus, the data suggested that mutations in this gene cause dominant RP, and that the encoded protein has an important but unknown role in photoreceptor biology.
Sullivan et al. (1999) likewise isolated the RP1 gene and identified mutations in affected families, including the R677X mutation in the UCLA-RP01 family as well as the Australian family (family D) previously studied by Xu et al. (1996), and a different nonsense mutation in the British family (UK-RP1) originally reported by Inglehearn et al. (1999) (Q679X; 603937.0004). They found that the 2 severely affected members of the UCLA-RP01 family were homozygous for RP1 mutations: 1 had noticeable night blindness at age 6, visual field loss by 8, and severe retinal atrophy and nonrecordable ERGs by age 18. Her youngest brother, examined at age 7, had experienced night blindness since early childhood and already had severe visual field constriction.
To determine the frequency and range of mutations in RP1, Bowne et al. (1999) screened probands from 56 large adRP families for mutations in the entire gene. After preliminary results indicated that mutations seemed to cluster in a 442-nucleotide segment of exon 4, an additional 194 probands with adRP and 409 probands with other degenerative retinal diseases were tested for mutations in this region alone. Bowne et al. (1999) identified 8 different disease-causing mutations, 6 of which were novel, in 17 of the 250 adRP probands tested. All of these mutations were either nonsense or frameshift mutations and led to severely truncated proteins. Based on this study, Bowne et al. (1999) estimated that mutations in RP1 cause at least 7% of adRP and that the 5-bp deletion (603937.0002) and the R677X mutation account for 59% of these mutations.
In all affected members of a large family segregating adRP linked to the RP1 locus (Iannaccone et al., 1996), Guillonneau et al. (1999) identified the R677X mutation (603937.0001); the mutation was absent in unaffected members and in 100 unrelated controls.
In affected members of 2 consanguineous Pakistani families with RP, Khaliq et al. (2005) identified homozygosity for a missense mutation in the RP1 gene (603937.0006). In affected members of another consanguineous Pakistani family with RP, they identified homozygosity for a 4-bp insertion in RP1 (603937.0007).
Audo et al. (2012) studied a French cohort consisting of 114 patients with autosomal dominant RP and found a prevalence of RP1 mutations of 5.3%, similar to the prevalence reported in other cohorts from the United States and the United Kingdom. Audo et al. (2012) stated that variable penetrance of the disease was observed in their cohort as well as in others, and that most patients with RP1 mutations show classic signs of RP with relatively preserved central vision and visual fields.
Animal ModelLiu et al. (2005) studied retinal development in Rp1 -/- mice and found that as early as postnatal day 7, these mice had already undergone significant molecular retinal changes in response to the Rp1 lesion. The molecular responses to the disruption of Rp1 changed dramatically during development and were distinct from responses to the disruption of the photoreceptor transcription factors Crx (602225), Pde6b (180072), and Nrl (162080). Using microarray analysis, Liu et al. (2005) found evidence that the JNK signaling cascades are specifically compromised in Rp1 -/- retinas and that Rp1 and JNK cascades play integral roles in photoreceptor development and maintenance.
HistoryAlthough Spence et al. (1977) found linkage between RP and amylase-2 (104650) on chromosome 1, later information indicated a computer error that, when corrected, left no linkage of RP and AMY2. However, suggestion of linkage to Rh remained (Cook, 1977). Heterogeneity of dominant retinitis pigmentosa was indicated by the existence of at least 1 family unlinked to chromosome 1 markers (Cook, 1977). Sparkes et al. (1979) presented additional data supporting the Rh-RP linkage, which still did not reach the level of proof, however. Field et al. (1980) found a summed lod score of 1.26 for RP and Rh at recombination fractions of 0.20 (male) and 0.40 (female). From analysis of all available data, Rao et al. (1979) found 'nonsignificant evidence of linkage' (lod score = 1.31). Heckenlively et al. (1982) found positive lod scores with Rh. Field et al. (1982) reported further studies of linkage with 29 markers. The largest lod score was 1.51 with Rh, with an estimated recombination fraction of 20% in males and 40% in females. All of these observations were made in a Los Angeles family known as UCLA-RP01. The phenotype in this family was later mapped to chromosome 8 by Blanton et al. (1991).