Retinitis Pigmentosa 17
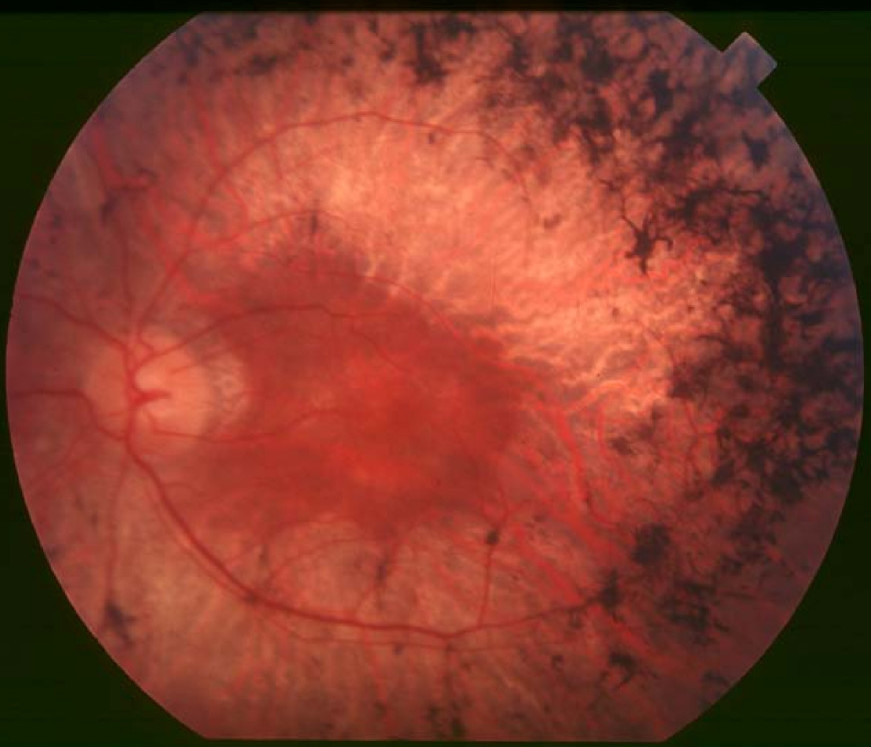
A number sign (#) is used with this entry because of evidence that retinitis pigmentosa-17 (RP17) is caused by heterozygous mutation in the gene encoding carbonic anhydrase IV (CA4; 114760) on chromosome 17q23.
For a phenotypic description and a discussion of genetic heterogeneity of retinitis pigmentosa, see 268000.
Clinical FeaturesYang et al. (2005) reported 13 affected individuals in a large Caucasian family with RP17. Reduction in night and peripheral vision manifested at around age 15, followed by rod photoreceptor atrophy, bone spicule pigmentation, and concomitant cone photoreceptor dysfunction manifested by photophobia, color vision changes, and decreased central vision. ERG demonstrated reduction of both rod and cone responses. Genealogic records indicated that this family was an offshoot of the South African family reported by Bardien et al. (1995).
MappingBardien et al. (1995) identified a form of autosomal dominant retinitis pigmentosa through linkage analysis with microsatellite markers in a large South African kindred. After exclusion of 13 RP candidate gene loci, including rhodopsin (RHO; 180380) and peripherin/RDS (PRPH2; 179605), they obtained positive lod scores at zero recombination for D17S808 (maximum lod = 4.63) and D17S807 (maximum lod = 5.69). Multipoint analysis gave a maximum lod score of 8.28 between these 2 markers. From haplotype analysis, the disease locus was found to lie in the interval between markers D17S809 and D17S942. Bardien et al. (1995) stated that the immigrant progenitor of the South African family in which they demonstrated linkage had numerous children and they studied only a single branch of the kindred. Further children of the initial immigrant also inherited the RP gene. Due to a founder effect, this mutation could be a common cause of autosomal dominant RP in South Africans of European descent.
Bardien et al. (1997) used a new series of microsatellite markers to localize the disease locus to 17q22. In addition, a second South African autosomal dominant RP family was shown to be linked to 17q22. Disease-associated haplotypes constructed for both families and multipoint linkage analysis placed the gene in a 10-cM interval between D17S1607 and D17S1874. Two candidate genes on 17q were excluded by finding recombination events between these genes and RP17: PDEG and TIMP2. Bardien-Kruger et al. (1999) studied an additional 17 members from the 2 unrelated South African families and refined the locus to a 1-cM interval between D17S1604 and D17S948.
In a large Dutch family with autosomal dominant RP, den Hollander et al. (1999) demonstrated linkage to the RP17 locus on 17q22 and refined the RP17 critical region to a 7.7-cM interval between markers D17S1607 and D17S948. Two positional candidate genes, AOC2 (602268) and GNGT2 (139391), were excluded by mutation analysis.
Molecular GeneticsIn affected members of 2 previously reported South African families with RP17 (Bardien et al., 1995; Bardien et al., 1997), Rebello et al. (2004) identified an R14W mutation (114760.0001) in the CA4 gene. The mutation was found to cosegregate with the disease phenotype in the 2 families and was not found in 36 unaffected family members or 100 controls.
In affected members of a European family with RP17, Yang et al. (2005) identified heterozygosity for an R219S mutation (114760.0002) in the CA4 gene.
In a screen of 96 Chinese patients with autosomal dominant RP, Alvarez et al. (2007) identified 1 patient with an R60H mutation (114760.0003) in the CA4 gene. They stated that functional analysis of this mutation provided further evidence that impaired pH regulation may underlie photoreceptor degeneration in RP17.