Chromosome 1p36 Deletion Syndrome
A number sign (#) is used with this entry because it represents a contiguous gene deletion syndrome caused by haploinsufficiency of a number of genes.
DescriptionThe constitutional deletion of chromosome 1p36 results in a syndrome with multiple congenital anomalies and mental retardation (Shapira et al., 1997). Monosomy 1p36 is the most common terminal deletion syndrome in humans, occurring in 1 in 5,000 births (Shaffer and Lupski, 2000; Heilstedt et al., 2003).
See also neurodevelopmental disorder with or without anomalies of the brain, eye, or heart (NEDBEH; 616975), which shows overlapping features and is caused by heterozygous mutation in the RERE gene (605226) on proximal chromosome 1p36.
Clinical FeaturesBedell et al. (1996) cited reports of 11 children who have been described with 2 or more syndromes with overlapping phenotypes and variations of the following features: short stature (9 of 10), prominent forehead (9 of 9), brachycephaly (7 of 7), microcephaly (10 of 11), midface hypoplasia (10 of 10), prominent jaw/chin (11 of 11), dysplastic pinna (6 of 7), hearing loss (3 of 11), congenital heart disease (7 of 11), hypotonia (11 of 11), DD/MR (11 of 11), facial clefting (4 of 11), and early demise (3 of 11). All patients were associated with a deletion of the terminal short arm of chromosome 1.
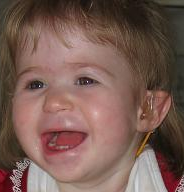
Based on the 13 subjects described by Shapira et al. (1997), facial characteristics of the syndrome include deep-set eyes, flat nasal bridge, asymmetric ears, and pointed chin. Additional clinical characteristics include seizures, cardiomyopathy, developmental delay, and hearing impairment (Slavotinek et al., 1999; Shaffer and Heilstedt, 2001).
Heilstedt et al. (2003) reported 62 patients with monosomy 1p36. Thirty were systematically examined through a specific protocol including hearing evaluations, palatal and ophthalmologic examinations, echocardiograms, neurologic assessments, and thyroid function tests. Orofacial clefting anomalies were present in 5 of 30 (17%); hypermetropia was present in 20 of 30 (67%). Six of 30 (20%) had hypothyroidism. All 30 had developmental delay and mental retardation. Twenty-six of 30 (87%) had hypotonia. Oropharyngeal dysphasia was present in 21 of 29 (72%). A history of dilated cardiomyopathy in infancy was present in 7 subjects (23%). In none did the condition worsen over time. Thirteen subjects (43%) had a structural heart defect, most frequently patent ductus arteriosus. Some hearing impairment was present in 82% of the subjects, being sensorineural type in almost all.
Tan et al. (2005) reported a 16-year-old boy with features of Cantu syndrome (239850) who was found to have a distal 1p36 deletion. The boy also had features not previously described in either syndrome, including hypercholesterolemia, type II diabetes, recurrent bony fractures, and nonalcoholic steatohepatitis.
Neal et al. (2006) reported a 3-year-old girl who had developmental delay, Duane syndrome anomaly, hearing loss, mild dysmorphic facial features including posteriorly rotated and slightly low-set ears and a broad nasal bridge, and scoliosis. MRI brain imaging revealed left periventricular nodular heterotopia (see PVNH1; 300049), truncation of the rostrum of the corpus callosum, slight ventricular enlargement, and patchy areas of hyperintensity consistent with delayed myelination. FISH analysis detected a deletion of 1p36 with loss of heterozygosity between D1S468 and D1S450, indicating at most a 9.6-Mb deletion region on 1pter-p36.22. Sequencing of the FLNA gene (300017), which has been shown to cause PVNH1, revealed no alterations in the coding region in this patient.
Battaglia et al. (2008) evaluated 60 patients with the 1p36 deletion syndrome (41 females and 19 males). Microcephaly was reported in 95% of patients, and all patients had straight eyebrows, deep-set eyes, midface hypoplasia, broad nasal root/bridge, long philtrum, and pointed chin. Other dysmorphic features included microbrachycephaly (65%), epicanthus (50%), large late-closing anterior fontanel (77%), and posteriorly rotated low-set abnormal ears (40%). Brachy/camptodactyly and short feet were prominent. Heart defects were present in 71%, including 23% with noncompaction cardiomyopathy. Other findings included visual inattentiveness (64%), visual abnormalities (52%), sensorineural deafness (28%), skeletal abnormalities (41%), abnormal genitalia (25%), and renal abnormalities (22%). Eighty-eight percent had central nervous system anomalies: 44% had seizures and 95% had hypotonia. All patients had developmental delay with poor or absent speech, and 47% had a behavior disorder. Gradual developmental progress was observed in all patients over time.
Rudnik-Schoneborn et al. (2008) reported an 8-month-old girl with microcephaly and a midline brain malformation who had an interstitial deletion of 1p36 on conventional chromosome analysis; FISH and array CGH analysis documented an 8.7-Mb deletion encompassing 1p36.23-p36.13. Brain MRI at age 5 months revealed agenesis of the anterior commissure and rostral corpus callosum and partial agenesis of the septum pellucidum. The authors stated that this structural brain defect had not previously been described in proximal 1p36 deletion.
Bursztejn et al. (2009) reported an 8-year-old girl with an initial clinical diagnosis of Aicardi syndrome (304050) who was subsequently found to carry a de novo 11.73-Mb terminal deletion of chromosome 1p36, thus revising the diagnosis. She had onset of infantile spasms at age 3 months, bilateral pupillary coloboma, agenesis of the corpus callosum, and delayed psychomotor development. Other features included deep-set eyes, low-set and posteriorly rotated ears, brachydactyly, and hypertrichosis. She also had interatrial and trabeculated interventricular communications. The deletion was found to occur on the maternal chromosome during oogenesis. The report emphasized the phenotypic overlap between the 2 disorders.
D'Angelo et al. (2010) described 9 unrelated patients with de novo deletions of distal 1p36 ranging in size from 2.2 to 10.2 Mb. Four deletions that could be studied occurred on the maternal allele. Four of the patients were ascertained from a larger group of 154 patients with psychomotor delay associated with hyperphagia and obesity, suggesting that this is an additional variable feature of monosomy 1p36. Five of the patients were ascertained from a larger group of 83 patients suspected to have monosomy 1p36 due to mental retardation. Three of the patients with obesity did not have the typical facial features of monosomy 1p36 and had slightly milder cognitive impairment. D'Angelo et al. (2010) suggested involvement of the PRKCZ gene (176982), which was deleted in all patients, but also noted the possibility of a position effect.
Dod et al. (2010) reported a 25-year-old man with monosomy 1p36 who developed symptoms of left ventricular noncompaction (LVNC; 604169) as an adult. In infancy and childhood, he had severe developmental delay, facial dysmorphism, seizures, and a cardiomyopathy with a low ejection fraction (15 to 20%). He also had scoliosis and spastic quadriparesis.
CytogeneticsHeilstedt et al. (2003) evaluated the deletion sizes in 61 subjects with monosomy 1p36 from 60 families using a contig of overlapping large-insert clones for the most distal 10.5 Mb of 1p36. They found pure terminal deletions, interstitial deletions, derivative chromosomes, and more complex rearrangements. Though some clustering of breakpoints was demonstrated, there was no single common breakpoint. They found that 60% of de novo 1p36 terminal deletions arose from the maternally inherited chromosome.
Heilstedt et al. (2003) ascertained 62 patients with deletions of 1p36 from 61 families. Most of the deletions occurred on the maternally derived chromosome. They identified terminal deletions, interstitial deletions, derivative chromosomes, and complex rearrangements. Retrospectively, 98% of deletions could be identified by routine chromosome analysis with careful attention to 1p36. Developmental delay/mental retardation was the most common clinical indication. Increased maternal serum alpha-fetoprotein (AFP; 104150) was detected in 4 of the 5 prenatally diagnosed cases. Maternal age at the time of birth of the affected child was significantly lower than the general United States population mean. The photograph of a patient demonstrated flat nasal bridge and nose, with pointed chin.
To further delineate genotype/phenotype correlations in monosomy 1p36, Redon et al. (2005) applied microarray-based comparative genomic hybridization, using an overlapping clone microarray covering 99.5% of the euchromatic portion of chromosome 1, to 6 patients with clinical features characteristic of monosomy 1p36. Deletions were confirmed in all patients. Two patients who displayed very similar features (facial characteristics and mental retardation) had distinct and nonoverlapping 1p36 deletions. Redon et al. (2005) suggested that the monosomy 1p36 syndrome may be due to a positional effect of the 1p36 rearrangement rather than haploinsufficiency of contiguous genes in the deleted region.
Monosomy 1p36 is characterized by marked variability in the size of the deletions, with no common breakpoints. In a review of the disorder, Gajecka et al. (2007) found no correlation between the deletion size and number of clinical features observed. An assessment of 1p36 deletions found that most (52 to 67%) were pure terminal deletions, followed by interstitial deletions (10 to 29%), unbalanced translocations (7 to 16%), and complex rearrangements (7 to 12%).
El-Hattab et al. (2010) described an infant girl with OEIS complex (258040) and chromosome 1p36 deletion who displayed features of both disorders, including omphalocele, cloacal exstrophy, imperforate anus, sacral segmentation defects, renal malposition and malrotation, genital anomalies, diastasis of the symphysis pubis, microbrachycephaly, large anterior fontanel, cardiac septal defects, rib fusion, limb deformity, developmental delay, and typical facial features. At 12 months of age, the patient developed bowel obstruction which progressed to septic shock and multiorgan failure and she died. Chromosomal microarray analysis detected a 2.4-Mb terminal deletion of chromosome 1p. There was no evidence of uniparental disomy. El-Hattab et al. (2010) suggested that OEIS complex might be caused by recessive mutation of a gene located in the 1p36 region, with the deletion uncovering a mutation located on the intact homolog; however, they noted that this was the first reported case of OEIS complex in association with a 1p36 deletion and that it was also possible that this case represented the chance occurrence of 2 independent conditions.
In 2 sisters who were originally described by Graham et al. (2004) and who exhibited features of Warburg Micro syndrome (see 600118), including microcephaly, cataract, microcornea, cortical blindness, increased frontal cortical thickness, enlarged ventricles, simplified irregular gyral pattern, generalized hypotonia, seizures, profound mental retardation, hypoplastic labia minora, hypertrichosis, and postnatal growth retardation, Handley et al. (2013) identified an unbalanced microdeletion/microduplication involving chromosomes 1p36 and 21q22. There was an approximately 6.9- to 7.1-Mb deletion from chromosome 1p36.33 to 1p36.23, containing the critical region for 1p36 deletion syndrome, as well as an approximately 5.6- to 6.0-Mb duplication from 21q22.3 to 21qter, distal to the Down syndrome (190685) critical region. Parental karyotyping confirmed that the sisters' father was a carrier of a balanced translocation. Genotyping of microsatellites covering the 1p36 deletion interval in both sisters revealed distinct maternal haplotypes, thus excluding the possibility that a new recessive gene was contributing to the phenotype.
DiagnosisHeilstedt et al. (2003) suggested a multistep approach in cases of monosomy 1p36 to give the most accurate counseling information: first, identification of the deletion of 1p36 by careful cytogenetic analysis and FISH with a probe containing the CDC2L1 locus (176873); second, telomere region-specific FISH to identify derivative chromosomes; and third, FISH using informative probes in the parents of those with the derivative chromosomes to uncover parental translocations (obviously significant to genetic counseling). Finally, the clinician's suspicion of a diagnosis of monosomy 1p36 is invaluable for leading to the correct diagnosis.
Prenatal Diagnosis
Campeau et al. (2008) described 2 patients with monosomy 1p36 syndrome who had brain abnormalities, particularly hydrocephalus and ventriculomegaly, detected by prenatal ultrasound. In 1 patient, amniocentesis and karyotyping did not show the deletion, which became apparent only by using array comparative genome hybridization. A review of previously reported cases showed that brain abnormalities are frequent, if not consistent, findings in this deletion syndrome. Such abnormalities include hydrocephalus, polymicrogyria, cerebral atrophy, and agenesis of the corpus callosum. Campeau et al. (2008) noted that certain small or interstitial deletions may not be identified by standard methods.
Molecular GeneticsBedell et al. (1996) further characterized the region of deletion in a patient with the karyotype 46,XY, del(1)(p36.3) and identified a gene that maps within the deleted region, the dishevelled-1 gene (DVL1; 601365). The authors speculated that this gene may play a role in the pathogenesis of the observed syndromes through haploinsufficiency or through genomic imprinting.
Windpassinger et al. (2002) mapped the gamma-aminobutyric acid A receptor delta subunit gene (GABRD; 137163) to chromosome 1p36.33, within the critical region of gene loss for the 1p36 deletion syndrome. As the gene encodes a GABA channel in the brain, the authors suggested that it may be a candidate for the neurodevelopmental and neuropsychiatric anomalies seen in the syndrome.
Rosenfeld et al. (2010) reported 5 patients with 200 to 823-kb overlapping interstitial deletions of chromosome 1p36.33 associated with classic features of the syndrome, including mental retardation, dysmorphic features, hypotonia, behavioral abnormalities, and seizures. The smallest region of overlap was 174 kb and encompassed 5 genes, 1 of which may be involved in signaling in neurons (GNB1; 139380). Two of the patients with seizures had deletions of GABRD, and 3 patients had deletions of PRKCZ (176982) and SKI (164780).
In 17 of 18 patients with a deletion in chromosome 1p36 who showed evidence of heart muscle disease, including left ventricular noncompaction (LVNC) or cardiomyopathy (see 615373), Arndt et al. (2013) aligned the regions of chromosomal loss and identified a shared deleted interval at chr1:3,224,674-3,354,772 bp (GRCh37) that involved only a single gene, PRDM16 (605557). Sequencing of PRDM16 in LVNC probands or posttransplantation patients with dilated cardiomyopathy identified 7 mutations (see, e.g., 605557.0001-605557.0006) that were not present in controls or exome sequencing databases. In 14 of the 18 patients with a deletion in chromosome 1p36, the SKI gene was also deleted; however, analysis of SKI in an independent LVNC cohort revealed no mutations.
De Leeuw and Houge (2014) stated that the results of their analysis of the evidence presented in the paper by Arndt et al. (2013) did not support the conclusions of those authors, and that it was unlikely that PRDM16 (605557) is a cause of cardiomyopathy in 1p36 deletion syndrome. Arndt et al. (2013) agreed that haploinsufficiency of PRDM16 is an unlikely or uncommon cause of cardiomyopathy in 1p36 deletion syndrome. However, they cited 3 additional independent lines of evidence supporting the role of PRDM16 in cardiomyopathy: first, PRDM16 mutations in nonsyndromic forms of left ventricular noncompaction (604169); second, a highly significant excess of deleterious PRDM16 variants in adult dilated cardiomyopathy (115200); and third, in vivo modeling data of several PRDM16 variants in zebrafish.