Retinitis Pigmentosa 43
A number sign (#) is used with this entry because this form of retinitis pigmentosa (RP43) is caused by homozygous or compound heterozygous mutation in the PDE6A gene (180071), encoding the alpha subunit of cGMP phosphodiesterase, on chromosome 5q31-q33.
For a phenotypic description and a discussion of genetic heterogeneity of retinitis pigmentosa (RP), see 268000.
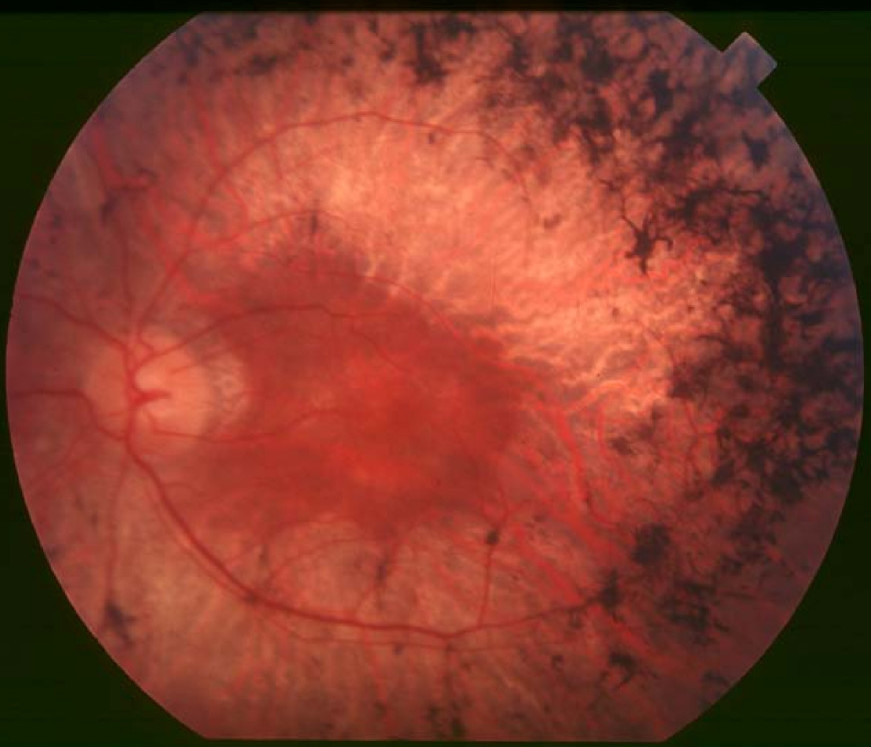
Clinical FeaturesHuang et al. (1995) studied 2 unrelated families segregating autosomal recessive retinitis pigmentosa (RP). Affected individuals reported night blindness since early childhood. Visual field testing revealed marked peripheral field loss. Fundus examination showed abnormalities typical of RP including waxy pallor of the optic discs, attenuated retinal vessels,
Corton et al. (2010) described a 5-generation consanguineous Spanish pedigree segregating autosomal recessive RP, in which 3 sibs and their nephew exhibited common symptoms such as bilateral vision impairment and night blindness from early childhood, abolished electroretinogram (ERG) responses, and fundus features typically associated with RP, including optic disc pallor, attenuated vessels, and intraretinal clumping of pigment. Unaffected members of the family had normal ophthalmologic examinations.
Molecular GeneticsBecause null mutations in the beta subunit of cGMP phosphodiesterase (PDE6B; 180072) cause some cases of retinitis pigmentosa (RP40; 613801), and since both alpha and beta subunits are required for full phosphodiesterase activity, Huang et al. (1995) examined the PDE6A gene (180071), encoding the alpha subunit, in 340 unrelated patients with RP. They found 3 point mutations in PDE6A (e.g., 180071.0001) in affected members of 2 pedigrees with recessive RP (RP43). Each mutation altered an essential functional domain of the encoded protein and likely disrupted its catalytic function.
In a 5-generation consanguineous Spanish pedigree segregating autosomal recessive RP, Corton et al. (2010) performed haplotype analysis using 88 SNPs of 22 known arRP genes and found consistent cosegregation for 3 genes; sequence analysis revealed homozygosity for a missense mutation in the PDE6A gene in affected individuals (V685M; 180071.0004) that was not found in 200 ethnically matched control chromosomes. The authors noted that the V685M variant is completely homologous to the mutation in the mouse model described by Sakamoto et al. (2009) involving nmf28/nmf28 mice, which showed an abnormal grainy-appearing fundus pigmentation with loss of nearly all photoreceptors shortly after birth.
Animal ModelBy N-ethyl-N-nitrosourea (ENU) mutagenesis screening in mouse, Sakamoto et al. (2009) identified missense mutations within the catalytic domain of the Pde6a gene. In these strains, significantly different biochemical outcomes and rates of degeneration of murine photoreceptor cells were observed, indicating allelic variation and previously unrecognized structure-function relationships. In addition, the mutations not only affected the function of the Pde6a protein itself, but also the level of Pde6b within the retina. Data suggested that the variation of the disease phenotype by background modifier genes may be dependent upon the particular disease allele present.