-
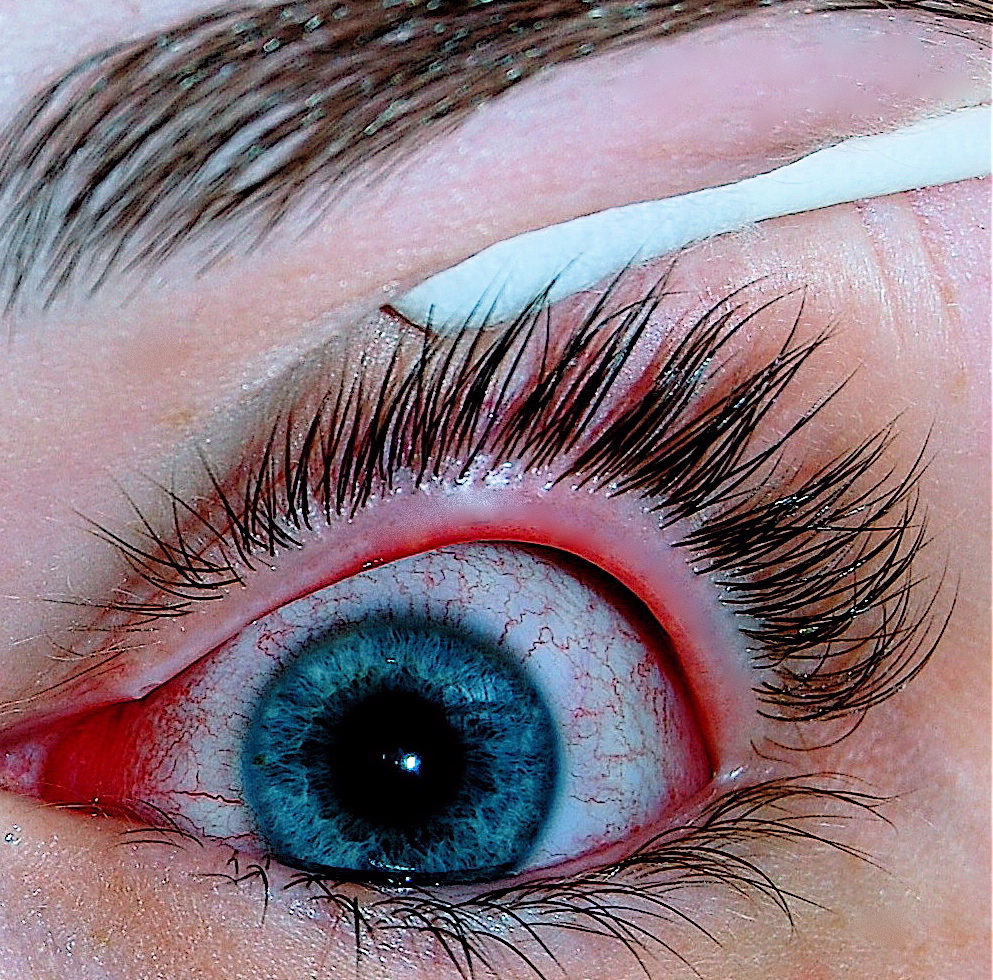
Conjunctivitis
Wikipedia
Vaccination against adenovirus, Haemophilus influenzae , pneumococcus, and Neisseria meningitidis is also effective. [28] Povidone-iodine eye solution has been found to prevent neonatal conjunctivitis. [29] It is becoming more commonly used globally because of its low cost. [29] Management [ edit ] Conjunctivitis resolves in 65% of cases without treatment, within 2–5 days. ... Immunology and Allergy Clinics of North America . 28 (1): 43–58, vi. doi : 10.1016/j.iac.2007.12.005 . ... Merck Manuals Professional Edition . Archived from the original on 28 December 2016 . Retrieved 31 December 2016 . ^ Hamborsky J, Kroger A, Wolfe C, eds. (2015).MMP2, PTAFR, PSMB9, LRRC8A, FERMT1, SAMD9, MMP1, MBTPS2, PIK3R1, PLG, POLH, PSMB4, PSMB8, RAG1, RAG2, ICOS, BLNK, TCF3, TNFRSF13B, GJB6, TNFRSF1A, UROS, WAS, WIPF1, XPA, XPC, TP63, STX16, SLC39A4, NLRC4, ERCC6, BTD, BTK, CD19, CD79A, CD79B, LRBA, COL7A1, CR2, DDB2, DKC1, DNASE1L3, USB1, ERCC2, ERF, FGFR2, FOXC2, GNAS, HLA-B, TNFRSF13C, IGHM, IGLL1, NLRP3, IKZF1, MBL3P, PLD3, IMMP1L, IGF2BP1, AD11, HM13, ALB, AOC3, IMPA1, CD14, GYPA, IFNGR1, IL4, CXCL8, IL10, IL12B, IL13, LGALS1, TYR, MBL2, PCOS1, ALK, CCL17, SOD3, STAT6, TEK, TNF, ERVK-19
-
Femoroacetabular Impingement
Wikipedia
Conservative treatment is often prescribed for those who have not yet received any therapy. [28] Conservative treatment includes physical therapy , avoidance of those activities that produce pain, and nonsteroidal anti-inflammatory drugs . [7] It may also include joint injections with cortisone or hyaluronic acid , particularly for those who wish to avoid surgery. [7] Physical therapy is implemented for the purpose of improving joint mobility, strengthening muscles surrounding the joint, correcting posture, and treating any other muscle or joint deficits that may be exacerbating the condition. [12] A movement analysis may also be performed to identify specific movement patterns that may be causing injury. [29] Studies to demonstrate the effectiveness of physical therapy are currently underway, with no conclusive results to date. [28] Operative treatment is generally recommended to those who continue to have symptoms. ... USCFootball.com . ^ "Hiljemark måste opereras - blir borta i tre månader" . fotbollskanalen (in Swedish) . Retrieved October 28, 2019 . ^ "Hip issue lands struggling Cardinals reliever Holland on DL" .
-
Lymphedema
Wikipedia
Bioimpedance measurement (which measures the amount of fluid in a limb) offers greater sensitivity than existing methods. [28] Chronic venous stasis changes can mimic early lymphedema, but the changes in venous stasis are more often bilateral and symmetric. ... PMID 9486670 . ^ Publishing, Licorn (2009-10-28). "Body image and quality of life in secondary lymphedema of the upper limb" . ... Scandinavian Journal of Plastic and Reconstructive Surgery and Hand Surgery . 28 (4): 289–93. doi : 10.3109/02844319409022014 .GATA2, FLT4, AKT1, PIEZO1, RASA2, RIT1, RRAS, SOS1, SOS2, TSC1, TSC2, VEGFC, LZTR1, IKBKG, DCHS1, ADAMTS3, MRAS, ABCC9, PTPN14, SLC35D1, KAT6B, LEMD3, SOX18, GJC2, ALG8, FAT4, SHANK3, ANTXR2, A2ML1, CCBE1, RAF1, PTEN, PTPN11, BCL2, BCL6, BRAF, CDC42, CDK5, EMP2, EPHB4, FOXC2, GLA, GUSB, HLA-DRB1, IGH, INPPL1, KIF11, KRAS, LBR, MPI, NAGA, NRAS, PIK3CA, PMM2, MAP2K1, MAP2K2, RELN, KIF7, HGF, MET, VEGFD
-
Ectopic Pregnancy
Wikipedia
Vaginal douching is thought by some to increase ectopic pregnancies. [23] Women exposed to DES in utero (also known as "DES daughters") also have an elevated risk of ectopic pregnancy. [28] However, DES has not been used since 1971 in the United States. [28] It has also been suggested that pathologic generation of nitric oxide through increased iNOS production may decrease tubal ciliary beats and smooth muscle contractions and thus affect embryo transport, which may consequently result in ectopic pregnancy. [29] The low socioeconomic status may be risk factors for ectopic pregnancy. [30] Diagnosis [ edit ] An ectopic pregnancy should be considered as the cause of abdominal pain or vaginal bleeding in every woman who has a positive pregnancy test . [1] The primary goal of diagnostic procedures in possible ectopic pregnancy is to triage according to risk rather than establishing pregnancy location. [4] Transvaginal ultrasonography [ edit ] An ultrasound showing a gestational sac with fetal heart in the fallopian tube has a very high specificity of ectopic pregnancy. ... A pregnancy not in the uterus. [31] The combination of a positive pregnancy test and the presence of what appears to be a normal intrauterine pregnancy does not exclude an ectopic pregnancy, since there may be either a heterotopic pregnancy or a " pseudosac ", which is a collection of within the endometrial cavity that may be seen in up to 20% of women. [4] A small amount of anechogenic -free fluid in the recto-uterine pouch is commonly found in both intrauterine and ectopic pregnancies. [4] The presence of echogenic fluid is estimated at between 28 and 56% of women with an ectopic pregnancy, and strongly indicates the presence of hemoperitoneum . [4] However, it does not necessarily result from tubal rupture but is commonly a result from leakage from the distal tubal opening . [4] As a rule of thumb, the finding of free fluid is significant if it reaches the fundus or is present in the vesico-uterine pouch . [4] A further marker of serious intra-abdominal bleeding is the presence of fluid in the hepatorenal recess of the subhepatic space . [4] Currently, Doppler ultrasonography is not considered to significantly contribute to the diagnosis of ectopic pregnancy. [4] A common misdiagnosis is of a normal intrauterine pregnancy is where the pregnancy is implanted laterally in an arcuate uterus , potentially being misdiagnosed as an interstitial pregnancy . [4] Ultrasonography and β-hCG [ edit ] Algorithm of the management of a pregnancy of unknown location, that is, a positive pregnancy test but no pregnancy is found on transvaginal ultrasonography . [4] If serum hCG at 0 hours is more than 1000 IU/L and there is no history suggestive of complete miscarriage, the ultrasonography should be repeated as soon as possible. [4] Where no intrauterine pregnancy (IUP) is seen on ultrasound, measuring β-human chorionic gonadotropin (β-hCG) levels may aid in the diagnosis. ... "Fertility after ectopic pregnancy: the DEMETER randomized trial" . Human Reproduction . 28 (5): 1247–53. doi : 10.1093/humrep/det037 .CRISP3, DNAAF2, CFAP298, TTC25, DNAL1, RSPH3, CFAP300, CCDC65, RSPH1, DRC1, CCDC114, LRRC56, CCDC151, DNAAF1, PIH1D3, DNAAF4, RSPH9, GAS2L2, CCDC39, MCIDAS, RSPH4A, DNAAF3, DNAJB13, DNAI2, CCDC103, DNAH5, DNAH1, GAS8, RPGR, SPAG1, OFD1, DNAH11, ARMC4, LRRC6, CCNO, DNAI1, STK36, NME8, ZMYND10, HYDIN, DNAAF5, CCDC40, VEGFA, TRO, MUC1, LIF, HOXA11, CNR1
-
Ehlers–danlos Syndromes
Wikipedia
This may progress to a life-threatening degree . [28] Heart conduction abnormalities have been found in those with hypermobility form of EDS. [29] Dilation and/or rupture ( aneurysm ) of ascending aorta [30] Cardiovascular autonomic dysfunction such as postural orthostatic tachycardia syndrome [31] [32] Raynaud's phenomenon Varicose veins [33] Heart murmur Heart conduction abnormalities Other manifestations [ edit ] Hiatal hernia [24] Gastroesophageal reflux [34] Poor gastrointestinal motility [35] Dysautonomia [36] Gorlin's sign (touch tongue to nose) [37] Anal prolapse [24] Flat feet Tracheobronchomalacia Collapsed lung ( spontaneous pneumothorax ) [13] Nerve disorders ( carpal tunnel syndrome , acroparesthesia , neuropathy , including small fiber neuropathy ) [38] Insensitivity to local anesthetics [39] Arnold–Chiari malformation [40] Platelet aggregation failure (platelets do not clump together properly) [41] Mast cell disorders (including mast cell activation syndrome and mastocytosis ) [42] Pregnancy complications : increased pain, mild to moderate peripartum bleeding, cervical insufficiency , uterine tearing, [15] or premature rupture of membranes [43] Hearing loss may occur in some types [44] Eye: Nearsightedness , retinal tearing and retinal detachment , keratoconus , blue sclera, dry eye, Sjogren's syndrome , lens subluxation, angioid streaks, epicanthal folds , strabismus , corneal scarring, brittle cornea syndrome, cataracts , carotid-cavernous sinus fistulas , macular degeneration [45] Craniocervical instability : caused by trauma(s) to the head and neck areas such as concussion and whiplash. ... Also, it can be misdiagnosed as EDS due to common symptoms, including fatigue, pain, gastrointestinal complaints, or cardiovascular autonomic dysfunction. [28] Osteoporosis and osteopenia are associated with EDS and symptomatic joint hypermobility [48] [49] Gorlin's sign in a case of EDS. ... It has no available genetic test. [28] Hypermobility EDS (hEDS) is the most common of the 19 types of connective tissue disorders. ... Archived from the original on February 28, 2009. ^ a b c d e f Chopra, Pradeep; Tinkle, Brad; Hamonet, Claude; Brock, Isabelle; Gompel, Anne; Bulbena, Antonio; Francomano, Clair (2017).COL1A1, COL1A2, COL3A1, B3GALT6, SLC39A13, DCN, PLOD1, COL5A1, B4GALT7, CHST14, TNXB, COL5A2, ADAMTS2, FN1, FKBP14, SEMA6A, HCRT, ADAMTS4, COX8A, CYP21A2, REM1, FLNA, DSE, AEBP1, TGFBR2, MAOA, TGFBR1, IL6, ELN, MED18, PDSS2, COL2A1, MMRN1, LRRK2, VWF, IL1R2, B3GALT4, CENPJ, ZEB2, TAB2, FAM20B, SEMA3A, CIT, COL5A3, SP140, TIMP2, ATP6V0A2, ACTA2, PAEP, TBX1, SNAI1, AR, ATP7A, BDNF, BGN, C1R, CGA, COL4A1, COL12A1, ATF2, CYP21A1P, DMPK, EDA, FBN1, MSTN, GDNF, IGFBP1, LRP5, NPPC, NPR2, OSM, ACTB, PDE4A, PLOD2, PTGS2, SLC6A3, EDS8
-
Gender Dysphoria In Children
Wikipedia
Following the ruling, NHS England announced that children under 16 would no longer be given puberty blockers without court authorization. [27] Prospective outcomes Gender dysphoria in children is more heavily linked to adult homosexuality than to an adult transgender identity, especially with regard to boys. [2] [3] [4] The majority of children diagnosed with gender dysphoria cease to desire to be the other sex by puberty , with most growing up to identify as gay, lesbian, or bisexual, with or without therapeutic intervention. [5] [6] [7] [28] Prospective studies indicate that this is the case for 60 to 80% of those who have entered adolescence ; puberty alleviates their gender dysphoria. [29] Bonifacio et al. state, "There is research to suggest, however, that [some desistance of GD] may be caused, in part, by an internalizing pressure to conform rather than a natural progression to non–gender variance." [30] If gender dysphoria persists during puberty, it is very likely permanent. [5] [7] For those with persisting or remitting gender dysphoria, the period between 10 and 13 years is crucial with regard to long-term gender identity. [29] Factors that are associated with gender dysphoria persisting through puberty include intensity of gender dysphoria, amount of cross-gendered behavior, and verbal identification with the desired/experienced gender (i.e. stating that they are a different gender rather than wish to be a different gender). [28] Prevalence The prevalence of gender dysphoria in children is unknown due to the absence of formal prevalence studies. [28] Gregor et al. state that "children who are not brought to the attention of specialised clinics do not feature in [gender dysphoria] studies and thus there may be a far greater prevalence of children with gender identity issues (who may or may not experience distress as a result) than these studies suggest." [31] Society and culture Diagnostic dispute Pickstone-Taylor has called Zucker and Bradley's therapeutic intervention "something disturbingly close to reparative therapy for homosexuals ." [32] Other academics, such as Maddux et. al , have also compared it to such therapy. [8] They argue that the goal is preventing a transgender identity because reparative therapy is believed to reduce the chances of adult GD, "which Zucker and Bradley characterize as undesirable." [33] Author Phyllis Burke wrote, "The diagnosis of GID in children, as supported by Zucker and Bradley, is simply child abuse." [34] Zucker dismisses Burke's book as "simplistic" and "not particularly illuminating;" and journalist Stephanie Wilkinson said Zucker characterized Burke's book as "the work of a journalist whose views shouldn't be put into the same camp as those of scientists like Richard Green or himself." [35] Critics argue that the GIDC diagnosis was a backdoor maneuver to replace homosexuality in the DSM; Zucker and Robert Spitzer counter that the GIDC inclusion was based on "expert consensus," which is "the same mechanism that led to the introduction of many new psychiatric diagnoses, including those for which systematic field trials were not available when the DSM-III was published." [36] Katherine Wilson of GID Reform Advocates stated: In the case of gender non-conforming children and adolescents, the GID criteria are significantly broader in scope in the DSM-IV (APA, 1994, p. 537) than in earlier revisions, to the concern of many civil libertarians. ... Retrieved August 11, 2018 . ^ "Gender incongruence (ICD-11)" . icd.who.int . WHO . Retrieved August 28, 2018 . ^ Alderson K (2012). Counseling LGBTI Clients . ... International Review of Psychiatry . 28 (1): 13–20. doi : 10.3109/09540261.2015.1115754 . ... "Gender Dysphoria in Children and Adolescents: A Review of Recent Research". Current Opinion in Psychiatry . 28 (6): 430–434. doi : 10.1097/YCO.0000000000000203 .
-
Chlamydia
Wikipedia
Some antibiotics such as β-lactams have been found to induce a persistent-like growth state. [28] [29] Diagnosis Chlamydia trachomatis inclusion bodies (brown) in a McCoy cell culture The diagnosis of genital chlamydial infections evolved rapidly from the 1990s through 2006. ... Centers for Disease Control and Prevention. December 28, 2009. Archived from the original on September 5, 2015 . ... Archived from the original on 2009-05-04 . Retrieved 2009-08-28 . ^ Desai, Monica; Woodhall, Sarah C; Nardone, Anthony; Burns, Fiona; Mercey, Danielle; Gilson, Richard (2015).IFNG, TNF, IL10, TLR4, NOD1, TLR2, NOS2, NECTIN1, IFNB1, MBL2, HLA-A, VDR, IL17A, TLR3, CASP9, MTA2, SHC1, CCV, TRBV20OR9-2, RAB11A, SLC18A1, SLC2A3, RIPK2, SLC2A1, TNFRSF10A, P2RX6, RAB6A, ACTB, ATG5, PIK3CG, MIR106A, RBM45, CYP2R1, CGAS, NLRP3, OTULIN, ARHGEF28, NOD2, TLR9, RAB14, ISYNA1, PYCARD, CLEC4E, P2RX2, ATF6, MMRN1, TREX1, PTGS2, PIGR, PIK3CD, CYP27B1, HSPD1, HLA-DRB1, FN1, ERN1, ENO1, SERPINB1, DHCR7, CRP, CXCL8, CCR7, CCR5, CD28, CASP3, C3, BCL2, STS, IL4, IL10RA, PIK3CB, P2RX5, PIK3CA, APAF1, FURIN, P4HB, P2RY2, P2RY1, P2RX7, P2RX4, IL18, P2RX3, P2RX1, MYD88, MIP, LGALS1, ITGB1, IDO1, P2RX5-TAX1BP3
-
Polysubstance Dependence
Wikipedia
It is a good idea that recovering addicts continue to attend social support groups or meet with counselors to ensure they do not relapse. [28] Inpatient treatment center [ edit ] Inpatient treatment centers are treatment centers where addicts move to the facility while they are undergoing treatment. ... Medications are a useful aid in treatments, but are not effective when they are the sole treatment method. [28] Medications that aid in treating addictions [ edit ] Methadone treatment for heroin addiction. [33] 'Naltrexone: Reduces opiates and alcohol cravings. ... Journal of the International Neuropsychological Society . 4 (4): 319–28. doi : 10.1017/S1355617798003191 .
-
Ethylene Glycol Poisoning
Wikipedia
As the metabolism of ethylene glycol progresses there will be less ethylene glycol and this will decrease the blood ethylene glycol concentration and the osmolal gap making this test less useful. [28] Additionally, the presence of other alcohols such as ethanol , isopropanol , or methanol or conditions such as alcoholic or diabetic ketoacidosis , lactic acidosis , or kidney failure may also produce an elevated osmolal gap leading to a false diagnosis. [7] Other laboratory abnormalities may suggest poisoning, especially the presence of a metabolic acidosis, particularly if it is characterized by a large anion gap . ... Poisoning of a raccoon was diagnosed in 2002 in Prince Edward Island, Canada. [70] An online veterinary manual provides information on lethal doses of ethylene glycol for chicken, cattle, as well as cats and dogs, adding that younger animals may be more susceptible. [71] History [ edit ] Ethylene glycol was once thought innocuous; in 1931 it was suggested as being suitable for use as a vehicle or solvent for injectable pharmaceutical preparations. [72] Numerous cases of poisoning have been reported since then, and it has been shown to be toxic to humans. [16] Environmental effects [ edit ] Ethylene glycol involved in aircraft de-icing and anti-icing operations is released onto land and eventually to waterways. [12] A report prepared for the World Health Organization in 2000 stated that laboratory tests exposing aquatic organisms to stream water receiving runoff from airports have shown toxic effects and death (p. 12). [73] Field studies in the vicinity of an airport have reported toxic signs consistent with ethylene glycol poisoning, fish kills, and reduced biodiversity, although those effects could not definitively be ascribed to ethylene glycol (p. 12). [73] The process of biodegrading of glycols also increases the risk to organisms, as oxygen levels become depleted in surface waters (p. 13). [73] Another study found the toxicity to aquatic and other organisms was relatively low, but the oxygen-depletion effect of biodegradation was more serious (p. 245). [74] Further, "Anaerobic biodegradation may also release relatively toxic byproducts such as acetaldehyde, ethanol, acetate, and methane (p. 245)." [74] In Canada, Environment Canada reports that "in recent years, management practices at Canada’s major airports have improved with the installation of new ethylene glycol application and mitigation facilities or improvements to existing ones." [12] Since 1994, federal airports must comply with the Glycol Guidelines of the Canadian Environmental Protection Act , monitoring and reporting on concentrations of glycols in surface water. [75] Detailed mitigation plans include storage and handling issues (p. 27), spill response procedures, and measures taken to reduce volumes of fluid (p. 28). [76] Considering factors such as the "seasonal nature of releases, ambient temperatures, metabolic rates and duration of exposure", Environment Canada stated in 2014 that "it is proposed that ethylene glycol is not entering the environment in a quantity or concentration or under conditions that have or may have an immediate or long-term harmful effect on the environment or its biological diversity". [12] In the U.S., airports are required to obtain stormwater discharge permits and ensure that wastes from deicing operations are properly collected and treated. [77] Large new airports may be required to collect 60 percent of aircraft deicing fluid after deicing. [77] Airports that discharge the collected aircraft deicing fluid directly to waters of the U.S. must also meet numeric discharge requirements for chemical oxygen demand. [77] A report in 2000 stated that ethylene glycol was becoming less popular for aircraft deicing in the U.S., due to its reporting requirements and adverse environmental impacts (p. 213), and noted a shift to the use of propylene glycol (p. ... "Methanol and ethylene glycol intoxication". Critical Care Clinics . 28 (4): 661–711. doi : 10.1016/j.ccc.2012.07.002 .
-
Angiogenesis
Wikipedia
One study in particular evaluated the effects of DII4 on tumor vascularity and growth. [28] In order for a tumor to grow and develop, it must have the proper vasculature. ... Journal of Applied Physiology . 97 (3): 1119–28. doi : 10.1152/japplphysiol.00035.2004 . ... Proceedings of the National Academy of Sciences . 116 (28): 14270–14279. doi : 10.1073/pnas.1905309116 .MTCO2P12, PIK3CA, TYMP, EGFR, FGF2, FLT1, HGF, COL18A1, HIF1A, CXCL8, MMP2, MMP9, COX2, ING4, SETD2, PECAM1, PIK3CB, BCL2, PIK3CD, PIK3CG, PTGS2, HPSE, STAT3, TEK, TGFB1, THBS1, TM7SF2, TP53, SCO2, VEGFA, CXCR4, NRP1, VPS51, KDR, ANGPT2, ANGPT1, ENG, DLL4, AKT1, PGF, PTEN, CXCL12, RECK, IL24, CD34, IL17A, MIR21, FN1, CCL2, CHI3L1, ERBB2, CCN2, TNF, SEMA3A, IGF1, CASP3, IL6, PLG, ITGAV, VASH2, IL1B, EGF, HSP90AA1, COL4A3, CXCL1, F3, MET, FGFR1, MIR126, EPHB2, FOXM1, NDRG1, ANTXR1, POSTN, IFNB1, CXCR2, SERPINF1, NOTCH3, EGFL7, NCL, MYC, RAC1, VASH1, RAF1, MDM2, RPS6KB1, LGALS1, CD248, CCL5, SDC1, JAK2, MIR204, AQP1, BSG, PROM1, ANGPTL4, DDX53, AGER, NES, CLEC14A, NELL1, EZH2, OXER1, PRSS55, CYP4Z1, PTN, ADM, FZD4, EPO, VN1R17P, NOTCH1, PTK2, GPR151, GPRC6A, ANPEP, TGFA, RNASE3, LGALS3, RET, SPZ1, IL32, TUG1, ACKR3, MMP14, MIR17HG, MRGPRX3, ARHGAP24, MRGPRX4, MTA1, ZEB1, SEMA3F, KLF5, TP73, LPAR3, IL1A, PKM, VWF, IFNG, AGT, DCN, AGTR1, ANG, SPP1, MAPK1, IL10, NRP2, APEX1, SEMA4D, LGR6, EIF4E, ITGAM, MRGPRX1, TXN, HSPA5, GPR166P, NTRK1, SERPINE1, CRISP2, APOA1, RUNX3, S100A4, CXCR6, IL1RN, CD44, LEP, CNMD, ESM1, IL2, SLC16A1, PCBP4, KRAS, LPAR2, RGS5, ITGA5, MIR378A, ITGB3, CASP9, CXCL5, IDO1, CXCL10, CSF2, ROBO4, PAG1, KDM3A, NRAS, YBX1, ANGPTL2, OSM, ADGRE2, CD274, TBK1, MCAT, ALDH1A1, ACTB, MAPK8, PFKFB3, MAPK3, NOX1, ARNT, PRKD1, BRMS1, PKD2, SRGN, PLAU, PPARG, DKK1, NOS3, JAG1, EGLN1, ROBO1, BRF2, MDK, REN, OPN1LW, BRS3, KDM5A, CCR2, TRIT1, SPHK1, NGF, ARL6IP5, CEACAM1, MVD, BGN, PPP1R12A, BDNF, ANGPT4, NFE2L2, CD93, CCN1, EGFL6, ADRA2B, CD163, MALAT1, FLT4, EFNA1, MIR93, CTHRC1, TIMP1, IL17F, THBS2, MTOR, MTDH, EDNRA, ADRA1A, GABPA, TGFBR2, GH1, EPAS1, EDN1, CREBBP, IGFBP3, FOXO3, AIMP1, EPHA3, EPHB4, PDLIM7, ETS1, ETV2, CYP4Z2P, DAB2IP, KLF4, MIR34A, EIF4G1, FGF13, MIR30D, FGFR3, GAB2, VEGFD, FOXC2, MIR107, EPHA1, CTSB, HSPG2, ADGRE5, VHL, RASAL2, GPR4, MIR135B, WNT2, TNC, CDK6, CTNNB1, STK11, SOX17, SLIT2, FBLN5, SSTR2, MIR17, HMOX1, CD276, CD68, GPR42, SP1, ADRB2, SPARC, GTF2B, RCAN1, ACVRL1, PRDX2, DHCR24, TFPI2, HMGB1, SSTR4, SRA1, PLXND1, TPX2, IQGAP1, NR1I2, ADGRA2, IKBKE, HAND1, MIR4530, STAB1, ZEB2, GDF15, RIPOR2, HEY1, CXCL14, DAPK2, BCAR1, KLK4, FJX1, NTN1, TM4SF5, SOCS6, APLN, RAMP2, LYVE1, ANGPTL1, SMR3B, NET1, GLCE, SPAG11B, AIFM1, RAPGEF3, YAP1, FST, IGF2BP3, TMSB10, TMX2-CTNND1, TXNIP, ST6GALNAC2, PTTG1, CALCRL, RALBP1, EHD1, WIF1, MIR6868, HDAC6, KLRC4-KLRK1, MMRN1, KLRK1, ZHX2, PER2, RASSF1, ARTN, ABCB6, DNM1L, PTP4A3, EDIL3, RCAN3, RAD50, PSME3, PROCR, SUCNR1, PPP1R16B, C1QTNF6, MIR638, MIR622, GPR119, PWAR1, LTO1, SEMA3D, RICTOR, ANKS4B, CXCL17, MACC1, MIR613, TBPL2, MIRLET7B, MIR130A, MIR143, LRG1, EGLN3, MIR146A, DNER, PANK2, SEMA6D, PDGFD, TRIM11, NECTIN4, ELOF1, KISS1R, IL1F10, COX4I2, UBASH3B, DCLK3, TSR2, IL33, CCDC34, PLXNA4, MIR145, MIR148A, PRPF31, MIR382, MIR451A, MIR410, MIR494, MIR495, MIR497, MIR519C, MIR503, MIR506, ECSCR, MIR612, POU5F1P3, CRNDE, POU5F1P4, SPAG11A, MIR542, HNP1, MIR196B, MIR190A, MIR323A, MIR192, MIR195, MIR19B1, MIR20A, MIR206, MIR210, MIR214, MIR221, MIR296, MIR299, MIR29B1, MIR29B2, MIR302A, MIR98, TNFSF12-TNFSF13, DHDDS, ADM2, MMRN2, FOXP3, PLCE1, VTA1, PIAS4, EMCN, TMEM8B, SIX4, MIR885, ADA2, P4HTM, XAF1, CEMP1, BCAS3, PINX1, EPN3, AGGF1, MIR874, F11R, SNIP1, IL22, NOC2L, FBXO22, AGO1, GREM1, TSPOAP1-AS1, DKK3, DKK2, IL37, IL17B, GIT1, MCTS1, CD24, ZNRD1, SMARCAL1, IL20, IMP3, LAPTM4B, MEG3, CCDC88A, HIVEP3, ACE2, NTN4, TRPV4, PROK2, PIEZO2, PBLD, GOLPH3, ADGRL4, DCLRE1C, STK33, CARD14, BHLHE41, SLC52A2, MCPH1, OPRPN, CXCL16, SUGP1, ADAMTS9, SUSD2, EIF5A2, TMPRSS4, MIR590, PAK6, CHPT1, AKR1B10, TRIB3, DANCR, NLGN2, SEMA6A, USP28, SCUBE2, DLK1, GALR2, ABR, MPDZ, GLRX, FPR2, FRG1, NR5A1, GCY, GDF2, GJA1, GLI2, CCR10, LAMA5, CXCR3, CXCL2, GRP, H2AX, HDAC1, HDAC2, HDGF, FPR1, FOSB, FOS, FOLH1, CTTN, ERBB4, ERN1, ESR1, ETS2, EYA1, F2, F2R, FABP5, PTK2B, EFEMP1, FGF7, FGFR4, FOXD1, FMOD, CFH, HIC1, HMGCS2, IGFBP7, ILF3, ILK, ITGA6, IRS1, ITGA9, ITGB5, JUN, JUNB, JUND, KISS1, KIT, KNG1, KIF11, KRT17, L1CAM, IL9, IGFBP6, HMGA1, IGFBP2, HOXB9, HRG, HSF1, HSPA1A, HSPA2, HSP90AB1, IARS1, ICAM1, ID1, IDH1, IDH2, IFN1@, IFNAR1, IFNAR2, IGF2, EMD, ELN, ELF4, ATF4, BAX, BMP2, BNIP3, BRAF, C4BPA, CA2, CAD, CAT, CAV1, RUNX2, RUNX1, SERPINH1, CCK, CD9, CD36, ATM, ARRB1, CDK4, AREG, ACVR1B, ADCYAP1R1, PARP1, AFP, AIF1, ALK, ALOX12, ALOX5, ALOX15, AMELX, ANXA2, ANXA5, ANXA6, FAS, AR, CD151, CDK5, ELF1, CTSL, DAB2, DECR1, CFD, DMPK, DNMT1, DUSP2, DUSP5, ECE1, ECM1, LPAR1, EFNA2, EFNA3, EFNB2, EGR1, EIF4EBP1, CYP2C9, CTNND1, CDKN1A, VCAN, CDKN2A, CEACAM5, CEBPD, CHGA, CMA1, CCR5, CCR6, COL2A1, COL11A2, KLF6, CRMP1, CRYAB, CSE1L, CSF1, CSF3, LAMA4, RPSA, ADAM15, TAZ, SMN1, SMN2, SST, STC1, TACR3, MAP3K7, TAT, ELOB, LCN2, TCF3, TEAD1, TEAD4, TFCP2, TFF3, TGFBR1, THBS4, SLPI, SLIT3, SLC22A3, SLC3A2, MOK, RAP2B, RARRES2, RBP3, TRIM27, RPL3, RPS15A, RPS24, RRAS, ACHE, S100A8, S100A13, CCL20, CCL22, SELE, TIMP3, TLR4, CLEC3B, SEMA3B, ADAM12, CDK2AP1, SLC7A5, MIA, PIK3R3, LGR5, DENR, TNFSF11, PSMG1, TP63, AKR1C3, NCOA1, EIF3C, EIF3I, TNFSF12, DEK, PAX8, NR2C2, PTP4A1, TRAF6, TRPC5, TSHR, TWIST1, TYR, SCGB1A1, UTRN, VEGFC, EZR, VLDLR, WNT5A, YY1, ZFP36, ZNF24, PCGF2, PXN, NECTIN2, PTX3, MMP1, MMP11, MMP12, ABCC1, MSH3, MT3, COX1, TRIM37, MYB, MYD88, CEACAM6, NCAM1, DRG1, NFIB, NGFR, NOS1, MMP7, MKI67, NOTCH2, CXCL9, LGALS8, LIFR, LOXL2, LPA, LTA, TM4SF1, SMAD2, SMAD3, SMAD4, MAS1, MCAM, MCL1, MDM4, MEF2D, MEFV, NOS2, NOVA2, PTHLH, PLD2, PLXNA1, POMC, POU5F1, PPARA, PPARD, PRKCD, MAP2K7, PRL, PRNP, LGMN, PSG4, PSMD4, PTGER2, PTGFR, PTGS1, PLRG1, PLAT, NPM1, PITX2, NR4A2, OLR1, OMP, PAEP, PRDX1, SERPINB2, PAK1, PCNA, PDC, PDGFB, PDGFRB, PF4, PGK1, SERPINA4, SERPINB5, CLIP1
-
Premenstrual Dysphoric Disorder
Wikipedia
Although there is a lack of consensus on the most efficient instrument by which to confirm a PMDD diagnosis, several validated scales for recording premenstrual symptoms include the Calendar of Premenstrual Experiences (COPE), Daily Record of Severity of Problems (DRSP), and Prospective Record of the Severity of Menstruation (PRISM). [26] [27] In the context of research, standardized numerical cutoffs are often applied to verify the diagnosis. [26] The difficulty of diagnosing PMDD is one reason that it can be challenging for lawyers to cite the disorder as a defence of crime, in the very rare cases where PMDD is allegedly associated with criminal violence. [28] DSM-5 [ edit ] The DSM-5 which established seven criteria (A through G) for the diagnosis of PMDD. [1] There is overlap between the criteria for PMDD in the DSM-5 and the criteria found in the Daily Record of Severity of Problems (DRSP). [26] [27] According to the DSM-5, a diagnosis of PMDD requires the presence of at least five of these symptoms with one of the symptoms being numbers 1-4. [1] These symptoms should occur during the week before menses and remit after initiation of menses. ... "Reproductive depression". Gynecological Endocrinology . 28 (s1): 42–45. doi : 10.3109/09513590.2012.651932 . ... "Estrogen-related mood disorders: reproductive life cycle factors". Advances in Nursing Science . 28 (4): 364–75. doi : 10.1097/00012272-200510000-00008 .
-
Echinococcosis
Wikipedia
People who undergo PAIR typically take albendazole or mebendazole from 7 days before the procedure until 28 days after the procedure. While open surgery still remains as the standard for cystic echinococcosis treatment, there have been a number of studies that suggest that PAIR with chemotherapy is more effective than surgery in terms of disease recurrence, and morbidity and mortality. [28] In addition to the three above mentioned treatments, there is currently research and studies looking at new treatment involving percutaneous thermal ablation (PTA) of the germinal layer in the cyst by means of a radiofrequency ablation device. ... Archived from the original on 4 December 2014 . Retrieved 28 November 2014 . ^ Vuitton, Dominique A.; McManus, Donald P.; Rogan, Michael T.; Romig, Thomas; Gottstein, Bruno; Naidich, Ariel; Tuxun, Tuerhongjiang; Wen, Hao; Menezes da Silva, Antonio; Vuitton, Dominique A.; McManus, Donald P.; Romig, Thomas; Rogan, Michael R.; Gottstein, Bruno; Menezes da Silva, Antonio; Wen, Hao; Naidich, Ariel; Tuxun, Tuerhongjiang; Avcioglu, Amza; Boufana, Belgees; Budke, Christine; Casulli, Adriano; Güven, Esin; Hillenbrand, Andreas; Jalousian, Fateme; Jemli, Mohamed Habib; Knapp, Jenny; Laatamna, Abdelkarim; Lahmar, Samia; Naidich, Ariel; Rogan, Michael T.; Sadjjadi, Seyed Mahmoud; Schmidberger, Julian; Amri, Manel; Bellanger, Anne-Pauline; Benazzouz, Sara; Brehm, Klaus; Hillenbrand, Andreas; Jalousian, Fateme; Kachani, Malika; Labsi, Moussa; Masala, Giovanna; Menezes da Silva, Antonio; Sadjjadi Seyed, Mahmoud; Soufli, Imene; Touil-Boukoffa, Chafia; Wang, Junhua; Zeyhle, Eberhard; Aji, Tuerganaili; Akhan, Okan; Bresson-Hadni, Solange; Dziri, Chadli; Gräter, Tilmann; Grüner, Beate; Haïf, Assia; Hillenbrand, Andreas; Koch, Stéphane; Rogan, Michael T.; Tamarozzi, Francesca; Tuxun, Tuerhongjiang; Giraudoux, Patrick; Torgerson, Paul; Vizcaychipi, Katherina; Xiao, Ning; Altintas, Nazmiye; Lin, Renyong; Millon, Laurence; Zhang, Wenbao; Achour, Karima; Fan, Haining; Junghanss, Thomas; Mantion, Georges A. (2020). ... Archived (PDF) from the original on 28 September 2011. ^ Bitton M, Kleiner-Baumgarten A, Peiser J, Barki Y, Sukenik S (February 1992). ... Besançon, March 27–29, 2014" . Parasite . 21 : 28. doi : 10.1051/parasite/2014024 .
-
Pneumonia
Wikipedia
Mixed infections with both viruses and bacteria may occur in roughly 45% of infections in children and in 15% of infections in adults. [12] A causative agent may not be isolated in about half of cases despite careful testing. [20] In an active population-based surveillance for community-acquired pneumonia requiring hospitalization in five hospitals in Chicago and Nashville from January 2010 through June 2012, 2259 patients were identified who had radiographic evidence of pneumonia and specimens that could be tested for the responsible pathogen. [28] Most patients (62%) had no detectable pathogens in their sample, and unexpectedly, respiratory viruses were detected more frequently than bacteria. [28] Specifically, 23% had one or more viruses, 11% had one or more bacteria, 3% had both bacterial and viral pathogens, and 1% had a fungal or mycobacterial infection. "The most common pathogens were human rhinovirus (in 9% of patients), influenza virus (in 6%), and Streptococcus pneumoniae (in 5%)." [28] The term pneumonia is sometimes more broadly applied to any condition resulting in inflammation of the lungs (caused for example by autoimmune diseases , chemical burns or drug reactions); however, this inflammation is more accurately referred to as pneumonitis . [15] [16] Factors that predispose to pneumonia include smoking , immunodeficiency , alcoholism , chronic obstructive pulmonary disease , sickle cell disease (SCD), asthma , chronic kidney disease , liver disease , and biological aging . [23] [29] [7] Additional risks in children include not being breastfed , exposure to cigarette smoke and other air pollution, malnutrition , and poverty. [30] The use of acid-suppressing medications – such as proton-pump inhibitors or H2 blockers – is associated with an increased risk of pneumonia. [31] Approximately 10% of people who require mechanical ventilation develop ventilator-associated pneumonia , [32] and people with a gastric feeding tube have an increased risk of developing aspiration pneumonia . [33] For people with certain variants of the FER gene , the risk of death is reduced in sepsis caused by pneumonia.IL1B, CXCL1, CCL2, CCL11, IKBKB, IL18, CCL17, ITGB2, TNF, TLR4, IL13, IL17A, IL6, IFNG, MYD88, IL33, TGFB1, HMOX1, IL4, CXCL5, SIRT1, CSF2, PARP1, CCL24, CXCR2, AHR, TNFRSF1A, CASP1, CD40LG, IL6ST, MYLK, AGT, SPP1, TNFRSF1B, EGR1, PF4, CCL8, CXCR3, CCL19, CCR2, POMC, IL1R2, CCR1, IL2RB, IL1R1, IL5RA, ABCF1, CCL4, CCL7, CCR6, CX3CL1, CCL20, CCR9, CXCL9, IL1A, ICAM1, CXCL2, IL1RN, MUC2, ADRB2, CFTR, CCL22, C3, ITGA4, SLC11A2, REG3A, ELANE, FER, FOXP3, SFTPC, NFKB1, SFTPA2, ICOS, CR2, PRKCD, CARD11, BTK, JAK3, ADA, RAG1, NFKB2, DNAL1, TTC25, DOCK8, SLC35A1, RAG2, IRF8, NPC2, HMGB1, IFNGR1, DNAH11, OFD1, ZAP70, WAS, SCGB1A1, TTR, IL2RG, TNFSF12, CFAP300, RSPH3, IL5, DNAAF4, PIH1D3, DNAAF1, CCDC151, TNFRSF13C, LRRC56, NLRP3, GAA, GABPA, GAS8, GFI1, CCDC114, DRC1, CCNO, TCIRG1, SLC9A6, UNC119, RSPH1, CCDC65, TNFRSF13B, CXCL8, LRRC6, DNAAF2, NPHS1, RYR1, CFAP298, NSMCE3, SERPINE1, RPGR, RMRP, SLC35C1, ARMC4, NFE2L2, CCDC40, NME8, RNF125, ZMYND10, DNAAF5, HYDIN, RANBP2, PTPRC, PNP, IL21R, TERT, SMARCD2, NIPBL, IL7R, DNAH1, IL10, STAT3, SRP54, SPAG1, DNAI1, STK36, SELENON, SGCG, LTBP3, DNAI2, SFTPD, DCLRE1C, MBL2, MPO, OSTM1, CHD7, LIG4, ZBTB24, DNMT3B, COPD, ACE, CD55, CCDC39, MCIDAS, RSPH4A, DNAAF3, DNAJB13, CD19, ACTA1, ACP5, CRP, CCDC103, MUC5B, ACADVL, CFB, GAS2L2, RSPH9, LRBA, CD81, CASP8, MS4A1, DNAH5, IL22, PPARG, ALB, MAPK14, ROS1, PARP9, GRAP2, POLDIP2, CRK, HIF1A, AIMP2, MAPK1, SLC27A5, DHFR, RNF19A, TLR2, CEL, AHSA1, PTGS2, PIK3CD, DHPS, DECR1, CXCL10, SFTPB, F2R, VEGFA, MIP, IL9, MYDGF, PIK3CG, IFNA13, COX2, SARS2, ALOX5, ANGPT1, IFNA1, BTBD8, CYBB, MMP9, AMBP, NM, SARS1, CD274, TREM1, CAV1, MIR155, BMP1, IL25, TIMP1, CCL5, IL17D, IFNB1, EGFR, PTGDS, ACE2, PRCP, IL2, SOCS3, MMP12, PGPEP1, PRMT1, MUC1, CD44, FOS, PDE4A, IL27, MTCO2P12, BRD4, TP53, PIK3CA, PIK3CB, ARG1, SIGIRR, LTB4R, SEMA7A, FCGR2A, CCR4, HPGDS, PTAFR, FOSB, NMU, NOD2, WDR26, MARCHF1, FOXM1, AGER, INSRR, BCL2, TRPM8, ITGAX, OSM, RAC1, NR1I2, JUN, JUNB, JUND, CLEC7A, S100A9, CD86, FPR1, IL37, VCAM1, ALOX15, ADAM8, TLR3, PLA2G1B, F2RL1, SMUG1, TSLP, CCN2, TFPI, RIPK3, ARIH1, F3, TAC1, CAMP, IL7, APOE, NOS2, PLG, HAMP, AKT1, MRC1, RGS2, CLEC1B, CD5L, PLAU, TSPO, CASP3, CD14, MAPK3, MIR223, HDAC6, IL23A, CENPJ, GDF15, RUNX1, PI3, CLOCK, HSPA14, ACSS2, PTPN11, PDCD1, TLR9, PROC, PLD2, CAT, MAPK8, SGPL1, RPS19, VIP, ANGPT2, THBS1, NKX2-1, TLR5, TM7SF2, TNFAIP6, ALOX12, NR2C2, ICOSLG, NBEAL2, TXN, KLHL2, PTGDR2, IRAK3, VTN, MIR511, TRPA1, HSPB3, VWF, CXCL13, CXCR4, ARHGEF5, AOC3, MASP2, ADORA2B, TNFRSF25, SORBS1, TIMELESS, PER2, LINC02605, PLA2G15, MAP3K7, ADAM17, PROCR, NAMPT, S100A12, PLXNC1, BDNF, MIR146B, CCL3, BCHE, SFTPA1, HACD1, AVP, ALDH7A1, ABCA3, STS, SDC1, SDC4, CXCL12, SELL, IL17B, AR, LPAR2, AQP5, IL17C, SLC12A2, UBAC1, CAP1, AD14, DEFB4B, HAVCR1, APOA1, PAEP, SERPINB6, P4HB, CHI3L1, CTNNB1, HP, CTAA1, HSPA1A, HSPA1B, HSPA4, HSPB1, HSPB2, HSPD1, ID2, CD276, CSF1, CCN1, CREBBP, IL2RA, CCR8, ZC3H12A, IL15, IL16, SLC52A2, MIR182, KNG1, LCN2, LEP, LGALS3, CTSG, ACCS, HAVCR2, IL22RA2, MLKL, ELN, MARK2, EMP1, PWAR1, HT, FABP4, FCGR1A, FCGR3A, DNASE1, SCGB3A2, FOXO3, HSH2D, FLT3LG, FPR2, UCN3, DEFB4A, GCLC, GPT, GPX1, GRN, NR3C1, LAMA1, GZMB, LNPEP, MALAT1, CYTB, CRLF2, NOTCH1, MMP13, NFATC2, MEFV, ACKR3, MCL1, MMP3, NOS3, MIR21, CD68, ENTPD1, NFKBIA, MUC5AC, MMP8, WWP2, HTRA3, IRAIN, IL17F, MIR20A, CLEC6A, LILRB4, BUD23, FSTL1, FBXO15, LCLAT1, TIRAP, IMMT, MORF4L1, JTB, LARP1BP2, RNU6-392P, NEAT1, NLRP12, CD248, ABHD6, MCEE, TUG1, SEMA6A, PES1, DUOXA1, PIGW, LOC102723996, CLEC9A, SLC52A1, SULF1, MPRIP, PSIP1, CEMIP, PHLPP2, HAGLROS, TPX2, CILK1, ESAM, OR10A4, POLG2, CYSLTR1, MTSS2, MIR200B, SEMA4A, TRAF3IP2, H3P42, PPM1K, FST, CBLL2, SPESP1, NOD1, NLRP6, KLF2, CCL26, RTN3, GSDMB, SPHK2, TICAM1, PGAM5, LRPPRC, ABCB6, LTB4R2, RBM8A, MRC2, PIKFYVE, TLK1, TRIM65, AREL1, PTPRVP, ATG7, GIPC1, TWIST2, RETNLB, SIX2, CXCR6, PGLYRP4, CELF2, POSTN, PDPN, DCD, IVNS1ABP, ARID3B, RNF19B, PRDX4, MIR200C, IL17RE, USE1, IL31RA, SIVA1, TRIM69, PBK, RLFP1, PIWIL4, PLEKHA7, BATF2, CFAP97, SPZ1, UBE2Z, IL20, TBX21, IL19, WNK4, COLEC11, CARD10, MIR17, MIR455, MMP28, MIR15A, MUL1, TNFAIP8L2, MIR636, TBK1, TUT7, PYCARD, MAP3K19, MIR150, DROSHA, CRPPA, C20orf181, WNT4, TRPV4, VSIR, MPPE1, ASCC2, MBL3P, RTEL1, MIR193A, P2RY12, MIR17HG, MIR326, NELFCD, ISYNA1, DCDC2, TRPV2, MIR370, PLF, TLR8, MIR197, BPIFA1, WG, HDAC9, GP6, AIDA, IFIH1, FXYD5, PINK1, IL20RA, CDK15, MIR146A, MIR199A1, MIR141, MIR127, MOXD1, MIR1247, IBTK, CRLS1, FEV, LINC01672, MIR27A, OCLN, OSBP2, NLRC4, SNHG16, MIR24-1, LY96, MICA, MIR23A, H3P44, MIR199A2, PADI4, JAM3, SEC14L2, EPG5, MIR34B, ITCH, SLC2A10, PTPN22, FGF21, IL31, CCDC22, STK39, MIR125A, MIR122, MIR100, MYO18A, RTL1, VIM2P, SCAF1, SIGLEC8, NAAA, EGLN1, CHIA, DIP, SND1, MIR29C, DNAJC5, SMOX, LATS2, SLC17A5, MIR1246, IL36G, PRSS55, ABCA1, SART3, GDF2, FOXD1, FLT3, FXN, MTOR, FUT2, ACKR1, GAS6, GATA3, GHRHR, FKBP4, GLA, GCLM, GOT2, GPR15, GSR, GSTM1, GSTP1, GSTT1, FOXF1, FGG, GZMA, F2, EPHX2, EPO, ERBB2, ERG, ESAT, ESR1, ESR2, ETS2, F8, FGF7, FABP5, FCAR, MS4A2, FCGR2B, FCGR3B, FCN2, FDPS, FES, GTF2H4, HCCS, PPP6R2, LCT, ITGA5, ITGAE, ITGAV, ITGB4, JAK1, KRT19, LAIR1, LAMC2, LGALS9, IRF5, LIF, LPA, LSP1, SH2D1A, NBR1, MAS1, MCAM, CD46, ISG20, IRF4, SERPIND1, NR4A1, CFH, CFHR1, HGF, HLA-C, HLA-DPB1, HLA-DQA2, HLA-DRB1, HLA-E, HPR, IDO1, HPX, HRG, HSD11B1, CFI, IFNAR1, IGF1, IL10RB, IL11, SERPINB1, EGF, EDNRA, BRCA1, BAAT, BAG1, BAX, BCL6, BDKRB1, CXCR5, BMP4, BMP6, BRCA2, ATM, BST1, SERPING1, C3AR1, C4BPB, C5AR1, CALCA, CD2, CD28, ATP4A, ATF3, EDN1, ANK1, AOC1, ADM, ADORA2A, ADORA3, GRK2, AGRP, AKT2, AKR1B1, ANXA2, ASAH1, ANXA5, AOAH, AOC2, APRT, FAS, FASLG, AQP1, AQP4, CD80, TNFRSF8, CD40, CYP2B6, CTLA4, CTSK, CTSS, CUX1, CYLD, CYP1A1, CYP1A2, CYP2A13, CYP2E1, CDC42, DDT, CFD, DPP4, DPT, HBEGF, ECE1, S1PR1, LPAR1, CST3, VCAN, CSF3, CPS1, CDKN2A, CEACAM5, CHRM3, CHRNA4, CHUK, CIRBP, CLCA1, CCR3, CCR7, CMKLR1, CNC2, CNR2, COL4A5, MAP3K8, COX8A, CP, CLDN7, MECP2, MEIS1, MFAP1, VASP, TRAF1, TRAF5, TRPM2, TTF1, TNFSF4, TYMS, UGCG, UROD, VCP, TNNI3, VDR, VHL, VIM, TRPV1, WNT5A, ZFP36, ST8SIA4, FOSL1, CRISP2, TNFAIP3, SLPI, TAGLN, SMPD2, FSCN1, SOD2, TRIM21, SSRP1, ST2, STAT1, SYK, TBXA2R, TIMP3, ZEB1, TRBV20OR9-2, PPP1R11, TEK, TFRC, TGFA, THBS2, KLF10, MIA, LOH19CR1, PLA2G6, CD83, CLDN1, SMC3, EXO1, SCAF11, ARHGEF2, XPR1, MSC, S1PR2, KLF4, FOXN1, SLIT2, ADIPOQ, KCNK6, ABCG2, AIM2, GGPS1, IL27RA, PTGES, CLDN2, GPRC5A, ARTN, MAP3K14, ST11, PIK3R3, IKBKG, PIAS1, SOCS1, PDE5A, BECN1, RNGTT, RIPK1, TNFSF14, TNFSF10, TNFRSF6B, TNFRSF11A, SOCS2, SPHK1, BTRC, USP13, SMPD1, SLC8A1, MIF, PLA2G4A, PDE7A, PDR, PECAM1, CFP, PGF, PHB, SERPINA1, PLA2G2A, PLA2G5, PCP4, PLAT, PLCL1, PLD1, POR, CTSA, PTPA, PREP, PRKACG, CDK16, PRKN, SLC5A1, NCF4, NR3C2, MMP2, CD200, MPP1, MSMB, COX1, MYH11, NCF2, NEO1, PEBP1, NFATC3, NINJ1, NPR1, NTF3, NTS, OAS3, P2RY1, P2RY6, PRKAR1A, PRKCA, PRSS2, CCL21, SAA2, SAA3P, SAFB, SAT1, ATXN7, SCT, CCL13, CCL18, SDC2, PSMD12, SELP, SELPLG, SETMAR, SFRP5, SGK1, SHBG, SHH, PMEL, SAA1, S100B, S100A1, RRBP1, PTGER3, PTGER4, PTGS1, PTX3, RAC2, MOK, RAP1B, RARA, RARB, PLAAT4, RASA1, REN, RENBP, RIT1, RNASE2, RNASE3, RORA, H3P10
-
Spondylolisthesis
Wikipedia
Plain Radiography (X-Ray) [ edit ] Plain radiography is often the first step in medical imaging. [17] Anteroposterior (front-back) and lateral (side) images are used to allow the physician to view the spine at multiple angles. [17] Oblique view are no longer recommended. [28] [29] In evaluating for spondylolithesis, plain radiographs provide information on the positioning and structural integrity of the spine. ... CS1 maint: multiple names: authors list ( link ) ^ Shamrock, Alan G.; Donnally III, Chester J.; Varacallo, Matthew (2019), "Lumbar Spondylolysis And Spondylolisthesis" , StatPearls , StatPearls Publishing, PMID 28846329 , retrieved 2019-10-28 ^ a b c d Tenny, Steven; Gillis, Christopher C. (2019), "Spondylolisthesis" , StatPearls , StatPearls Publishing, PMID 28613518 , retrieved 2019-10-28 ^ Frank Gaillard.
-
Personality Disorder
Wikipedia
People with borderline personality disorder have a pervasive pattern of instability in interpersonal relationships. [28] [ unreliable medical source? ] Histrionic Hysteria, dramatic, seductive, shallow, egocentric, attention-seeking, vain. ... DSM-III-R personality disorder diagnostic co-occurrence aggregated across six research sites [40] Type of Personality Disorder PPD SzPD StPD ASPD BPD HPD NPD AvPD DPD OCPD PAPD Paranoid (PPD) — 8 19 15 41 28 26 44 23 21 30 Schizoid (SzPD) 38 — 39 8 22 8 22 55 11 20 9 Schizotypal (StPD) 43 32 — 19 4 17 26 68 34 19 18 Antisocial (ASPD) 30 8 15 — 59 39 40 25 19 9 29 Borderline (BPD) 31 6 16 23 — 30 19 39 36 12 21 Histrionic (HPD) 29 2 7 17 41 — 40 21 28 13 25 Narcissistic (NPD) 41 12 18 25 38 60 — 32 24 21 38 Avoidant (AvPD) 33 15 22 11 39 16 15 — 43 16 19 Dependent (DPD) 26 3 16 16 48 24 14 57 — 15 22 Obsessive-Compulsive (OCPD) 31 10 11 4 25 21 19 37 27 — 23 Passive-Aggressive (PAPD) 39 6 12 25 44 36 39 41 34 23 — Sites used DSM-III-R criterion sets.SLC6A4, DRD2, GABRB3, MTA2, CHD2, SCN1A, MAPK10, KCTD17, SPG21, DNM1, CBS, CUX2, XK, TOR1A, SGCE, MAOA, SFSWAP, ITSN2, TPH2, SNAP25, PRL, WASF2, BRAP, ZNF185, CPSF4, THAS, SOX4, SPOCK3, KCNIP4, AMPD1, NHS, NRAS, LMNA, CHRNA7, COMT, CRP, CYP2D6, DPYD, DRD3, DRD4, TSC22D3, FGF9, NR3C1, HTR1A, HTR1B, HTR2A, IL6, IMPA1, DDX53
-
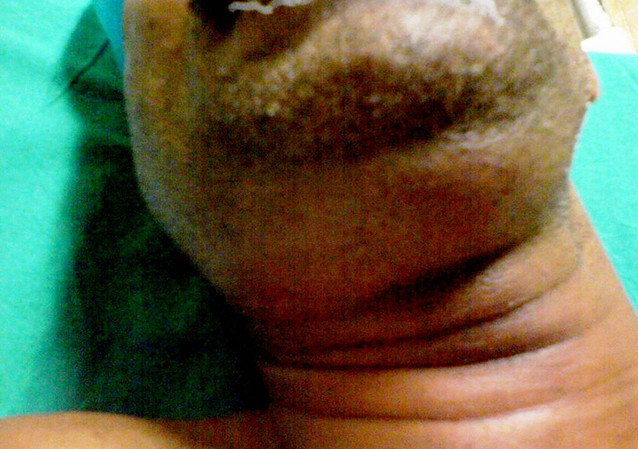
Ludwig's Angina
Wikipedia
Patients must therefore be well-nourished and hydrated to promote wound healing and to fight off infection. [28] Post-operative care [ edit ] Extubation , which is the removal of endotracheal tube to liberate the patient from mechanical ventilation, should only be done when the patient's airway is proved to be patent, allowing adequate breathing. This is indicated by a decrease in swelling and patient's capability of breathing adequately around an uncuffed endotracheal tube with the lumen blocked. [28] During the hospital stay, patient's condition will be closely monitored by: carrying out cultures and sensitivity tests to decide if any changes need to be made to patient's antibiotic course observing patient's body temperature - a rise implies further infection monitoring patient's white blood cell count - a decrease implies effective and sufficient drainage repeating CT scans to prove patient's restored health status or if infection extends, the anatomical areas that are affected. [28] Moreover, it is advised to never leave young children with significant neck swelling unattended and they should always be seated to prevent suffocation. [24] Etymology [ edit ] The term “angina”, is derived from the Latin word “ angere ”, which means “choke”; and the Greek word “ ankhone ”, which means “strangle”.
-
Huntington's Disease
Wikipedia
.), known as a trinucleotide repeat. [20] CAG is the three-letter genetic code ( codon ) for the amino acid glutamine , so a series of them results in the production of a chain of glutamine known as a polyglutamine tract (or polyQ tract), and the repeated part of the gene, the PolyQ region . [28] Graphic showing at top normal range of repeats, and disease-causing range of repeats. ... This probability is sex-independent. [31] Trinucleotide CAG repeats over 28 are unstable during replication , and this instability increases with the number of repeats present. [20] This usually leads to new expansions as generations pass ( dynamic mutations ) instead of reproducing an exact copy of the trinucleotide repeat. [20] This causes the number of repeats to change in successive generations, such that an unaffected parent with an "intermediate" number of repeats (28–35), or "reduced penetrance" (36–40), may pass on a copy of the gene with an increase in the number of repeats that produces fully penetrant HD. [20] Such increases in the number of repeats (and hence earlier age of onset and severity of disease) in successive generations is known as genetic anticipation . [1] Instability is greater in spermatogenesis than oogenesis ; [20] maternally inherited alleles are usually of a similar repeat length, whereas paternally inherited ones have a higher chance of increasing in length. [20] [32] It is rare for Huntington's disease to be caused by a new mutation , where neither parent has over 36 CAG repeats. [33] In the rare situations where both parents have an expanded HD gene, the risk increases to 75%, and when either parent has two expanded copies, the risk is 100% (all children will be affected).HTT, CNR1, OGG1, GDNF, SLC2A3, MAOA, AIFM1, SLC29A1, FAAH, RCAN1, DIABLO, PPARGC1A, PPP1R1B, MAOB, IP6K2, GAP43, NRSN1, GAL, NFE2L2, RELN, CNTF, ZDHHC17, CREB1, NPY, LEP, ITPR1, MMP9, PPARD, ZDHHC13, CNR2, BCL2, DRD1, DUSP1, PTGS2, GSR, ABAT, BAX, HAAO, QPRT, NOS3, E2F1, ND3, MBP, CAT, ADD1, FAN1, RNF4, ADGB, SH3BP2, SIRT1, REST, RGS12, MSANTD1, TP53, ATXN1, PDE10A, ATXN3, CASP6, GABPA, IGF1, SNCA, SOD1, JPH3, EPO, TGM2, SIGMAR1, GRK4, TBP, IL6, GRIN2B, HTT-AS, APOE, ALB, BDNF, HAP1, AR, CRP, DRD2, DNM1L, FXN, GFAP, SLC6A4, GRIK2, CREBBP, HSF1, CTNNB1, GAPDH, SIRT2, ACE, TNF, DNAJB6, EIF4E, ZNF141, PENK, OPN1LW, PTH, PAG1, IGFALS, CDK5, ATM, CSF2, MAPK1, PCBP4, MAPT, SLC1A2, NEFL, SMUG1, C9orf72, HSPA4, IFNL3, HIP1, NEAT1, FGF23, HCRT, SOST, GRM5, IL1B, LAMC2, HDAC4, SETDB1, NTRK2, IL10, TAC1, CASP3, PPARG, KMO, CALB1, ATXN2, PPP1R2C, GRIN2A, F8A1, DAPK2, LINC01672, TLR4, HDAC9, F8A3, ADORA2A, F8A2, DENR, HSP90AA1, MSH3, AKT1, SQSTM1, VEGFA, MSH2, PON1, PTX3, CRMP1, SIRT3, UBB, IL12B, UTRN, H2AX, HAMP, ACTB, ATF6, CXCL8, IL12A, GCG, FOXO3, TAL1, SLC18A2, ANGPT1, LY6E, IDO1, FUS, SCLY, KRT7, CCL5, MLH1, ATXN7, HIPK3, HDAC1, OPTN, SST, PPP1R11, TSHZ1, SRSF6, AGER, APP, STAT3, GPT, ACBD3, CCL2, BCL11B, FOXO1, CDKL1, TCERG1, UCHL1, BRCA1, DLG4, CDK20, PDE4A, TARDBP, ABCB1, KCNH4, PIK3CA, PIK3CD, PIK3CG, NPEPPS, UBE3A, NUP43, KCNH8, CACNA1A, CKB, CHI3L1, VCP, RCOR1, PRL, VRK2, PROS1, NT5C2, PSMC6, CASP1, H3P30, TM7SF2, PIK3CB, ELK1, MTDH, EPHB2, HDAC3, PTPN5, TGM1, NRF1, BDNF-AS, FAP, HYPK, PRKCA, MAPK11, AHSG, ADA, MGP, PRKAB1, MCF2L, KHDRBS1, MAP3K5, RGS2, SHBG, FOXP1, MFAP1, MOK, CXCR6, P4HB, PVALB, MAP2, DCTN4, ADCYAP1, TPPP, PHGDH, LGALS3, NPY2R, ALPP, TREM2, RAB11A, LRP2, SLC1A1, NGFR, NGF, PRNP, ALOX5, EIF2AK2, NTF4, PRKAA1, MMP2, MTHFR, ACP3, TUSC2, NCAM1, PYCARD, RASD2, CYP46A1, S100A12, FAM193A, AGT, MRPS30, SGSM3, TNFRSF11B, NUP62, REG3A, REN, SERPINE1, ADRA1A, PSAT1, UBQLN1, RXRA, PRKAA2, PDAP1, BECN1, FIS1, IFNL4, MRC1, NF2, PLAG1, AGO2, PIN1, PAPOLA, ASAP1, KCNIP3, ADRA2B, CCHCR1, ASAP2, CALB2, SLC2A1, DECR1, TNNI3, SLCO6A1, ATN1, TIMP2, LPAR2, TIMP1, EDNRA, EGF, EGR1, GRIN3A, EPHA3, UBXN11, TGFB1, TFRC, FCGR3A, FCGR3B, FEN1, FGF9, FGFR3, FRAXE, NR5A1, KALRN, SELENBP1, MSTN, MFN2, BRS3, TRPC5, GLO1, RLS1, CASP2, CASP8, CBS, VWF, CD28, MIR155, KL, ADIPOQ, MIR132, VDR, MIR10B, GPR52, ENHO, GSTK1, MIR34A, CP, TSPO, UBC, TYMS, TWIST1, CCN2, USP12, CUX1, CX3CR1, TTR, METRN, HTR1A, IAPP, ACKR3, KLK3, STIM2, IFNG, KIDINS220, HMOX1, APRT, HP, TOMM40, GTF2H1, SOD2, HSPD1, MEG3, ZBTB16, ATF7IP, APEX1, APAF1, MTPAP, HACE1, IFNA2, GSK3B, GPR42, ACE2, SPHK1, ITPR3, IFT57, SSTR4, NGB, ATG7, IER3, SPON2, HOMER1, CPQ, GDF15, ATF5, NLRP1, LIN7A, TUBB4B, TRIP10, SPTLC1, RPH3A, UBD, GSTO1, AHSA1, NREP, CD163, CCT2, CAP1, SORBS1, KLF4, HAND1, MLXIP, GRAP2, CIB1, PRMT5, APLN, SEMA3A, CERS1, PSIP1, PDIA6, CDK5R1, WDR5, ARFGEF2, BCL10, SOCS3, WWP1, IMMT, WASHC5, NR1H3, SV2B, SV2A, SBNO2, BTRC, CHEK2, CCL27, PNKP, BCAR1, APC2, MGLL, BAG3, MACROH2A1, CCL4L2, HDAC6, SGPL1, LZTS3, GABBR2, CLOCK, CPLX2, HACD1, DNAJA2, PSME3, KEAP1, NAT2, SV2C, MARCHF8, IL34, HJV, PDIK1L, ERFE, ZDHHC15, OXER1, PTCRA, SCAMP5, GPRC6A, EHMT1, STH, IPMK, GPX6, MRGPRX1, OR10A4, GOLGA8G, RAB7B, MCIDAS, CFAP54, GPR151, PPARGC1B, CD200R1, ASRGL1, TET1, UBXN6, SLC25A28, MAGT1, EVA1A, CYSTM1, DNAJC14, DCLK3, FOXP2, OPN4, NLRP3, CARD16, MRGPRX3, MRGPRX4, GSTO2, OSR1, NPSR1, CCL4L1, CIMT, POU5F1P3, POU5F1P4, MIR421, MIR549A, MIR615, CASC8, PSS, ZGLP1, MIR942, TIMM23, MIR1247, ECT, TUNAR, TMX2-CTNND1, MIR4488, GDNF-AS1, CBSL, LINC02605, MT1IP, MIR486-1, MIR106A, MIR146B, MIR122, MIR128-1, MIR146A, MIR150, MIR200A, MIR200C, MIR21, MIR214, MIR22, MIR222, MIR26B, MIR27A, MIR302A, MIR34B, VN1R17P, GPR166P, MIR196B, PANK2, RTL10, TBC1D9, GIT1, NSG1, NAAA, IL37, DISC1, PDLIM3, HPGDS, HTRA2, SLCO1B3, FLVCR1, MTHFSD, SETD2, DROSHA, MYLIP, UBQLN2, PACSIN1, SLC40A1, CHCHD2, AADAT, SIGLEC7, MLH3, AKAP8L, FGF21, ACSBG1, RRS1, PHLPP1, CRTC1, DICER1, ABCB10, BRD4, LPAR3, ARFIP2, IL17RA, QPCT, CHRDL2, RNF19A, SOSTDC1, NGDN, ATRNL1, POLDIP2, HSPA14, UBR5, NCKIPSD, CMTR2, GPRC5C, ZNF395, SPHK2, UBQLN4, SLC12A9, S100A14, SLC12A5, SORCS2, CFAP97, GBA2, HES4, CTDSP1, LGR6, PAPPA2, SLC39A8, LIN7B, ZMAT3, HDAC8, PRPF40A, SIRT6, GPRC5D, NUB1, TNFRSF6B, HDL3, S1PR5, IMMTP1, DDIT4, UGT1A1, TMEM106B, TMED9, AHI1, TUG1, ATG16L1, MAP1S, SBNO1, ATAD3A, NAT10, FBXW7, GHRL, CXCL12, RIPK2, DRD5, GATA4, GC, GH1, GIP, GJA1, GLP1R, GPR3, GPX4, GPX7, GRN, GRIK5, GRIN1, GRINA, GRM2, CXCL3, GYS1, GZMB, HCLS1, HDAC2, GAD2, GABRA2, XRCC6, F13B, DUSP2, E2F2, EMP1, ERCC6, ERN1, EZH1, F11, F13A1, FABP4, FSHMD1A, PTK2B, FBN1, FGF2, FGF13, FGFR2, FLNA, FMR1, MTOR, HIF1A, UBE2K, HLA-C, LBP, IL18, ITGA2B, JAG2, KCNA1, KCNA5, KCNA6, KDR, KPNA1, LBR, IL13, LCN2, LIMK1, LMNA, LNPEP, LPA, LPL, LTB, SMAD3, IL17A, IL9, HLA-DPB1, TNC, HMGB1, HNF4A, HPCA, HRES1, DNAJB2, DNAJA1, HSPA8, DNAJB1, IDH1, IL7, IDUA, IFNA1, IFNA13, IKBKB, IL1A, IL2RB, IL4, IL4R, DTX1, DRD3, IRS2, DPYSL2, ATP2A2, ALDH7A1, ATRX, B2M, BAG1, BCHE, BCL2L1, TNFRSF17, BGLAP, BNIP3, C3, C5, C5AR1, CAD, CALD1, CAMK2A, CAST, CAV1, CD14, ATHS, ATF4, STS, ADCYAP1R1, ABCA1, ASIC1, ACHE, ACO1, ACO2, ACVR2B, ACVRL1, ADAM10, ADM, AQP9, ADORA2B, PARP1, ABCD1, ALPI, ANGPT2, ANK1, ANXA3, XIAP, CD34, CD40, CD47, DCT, CSH2, CTAA1, CTNND1, CYBB, CYP3A4, DAB2, DAXX, DBH, DDIT3, CSF1R, DHCR24, DHFR, DLG1, DLG3, DMD, DNAH6, DNASE1, DPP6, CSH1, CSE1L, CDK9, CNTN1, CDKN1A, CDKN1B, CDKN2A, CDX2, CETP, CHAT, CHRM1, CLCN1, COL4A2, MAPK14, COL11A2, COMT, KLF6, ATF2, CRH, CRK, CRYAB, CS, MBL2, MC1R, MCL1, MECP2, SLCO2A1, SLPI, SUMO3, SUMO2, SNAP25, SNCB, SNCG, SORL1, SOX2, SOX4, SP1, SPN, SPP1, SPTBN2, SRI, STAT5A, STAT5B, STATH, SUPT4H1, SLC12A2, SLC12A1, SLC9A5, MAP2K4, RPS6KA1, RPS6KB1, RPS19, RRAS, SORT1, SCN4A, CCL4, SELENOP, SGCA, SLC9A1, SGK1, SH3GL3, SKIL, SKP1, SLC1A3, SLC1A6, SLC2A4, SLC6A3, SYN1, TAF4, TARBP2, ULK1, AIMP2, COIL, MIA, KDM5C, KDM5D, BAP1, FZD4, LOH19CR1, CUL1, RAB7A, DGKE, BLZF1, PIAS1, KHSRP, PSMG1, HSD17B6, AOC3, PDE5A, ST8SIA4, XPC, TCF3, TRPC1, TFAM, TGM3, TH, NKX2-1, CLDN5, TNFRSF1B, TRAF6, TRP-AGG2-6, TRPC4, XBP1, TXNRD1, SUMO1, UCP1, UCP2, KDM6A, VCL, VIP, VIPR2, ROS1, ROCK1, RNASE4, NFATC4, MTRR, MUTYH, PPP1R12A, NAIP, NDUFA1, NEFH, NEK1, NEUROD1, NNAT, MTNR1A, NOS1, NOS2, PNP, NPC1, NPM1, NPPA, NPY5R, SLC11A2, MTR, CYTB, NT5E, MSX1, MAP3K1, KITLG, MLF1, FOXO4, MMP14, MNAT1, CD200, MSN, MT1A, MT1X, MT1B, MT1E, MT1F, MT1G, MT1H, MT1JP, MT1M, MT1L, NRGN, NTF3, RLN2, PTMA, MAPK8, MAPK10, MAP2K1, MAP2K2, MAP2K6, PSEN1, PSG5, PTBP1, PTMAP4, PKIB, PTPN13, RAC1, RAF1, RAN, RANBP2, RARB, RENBP, RET, PKN2, PPT1, OPA1, PCP4, P2RX7, PAEP, PAM, PRKN, PC, PCBP1, PCM1, PCNA, PDE1B, POU5F1, PDK3, PEX7, PF4, PGD, SERPINB6, PITX3, PLCG1, PMS2, H3P10
-
Fabry Disease
Omim
-
Search Fabry disease case abstracts
Due to the high frequency of Fabry disease (4%, 28 of 721) in this cohort of stroke patients with cryptogenic stroke aged between 18 and 55 years, Rolfs et al. (2005) suggested that Fabry disease should be considered in young patients with unexplained stroke. ... Six of 32 women had renal failure, 9 of 32 (28%) died of cerebrovascular complications, and 42 (70%) had experienced neuropathic pain. ... Using a personality inventory questionnaire to evaluate the psychologic profile of 28 adult patients with Fabry disease, Crosbie et al. (2009) found that the patients had significantly higher (abnormal) scores on several scales, including hypochondriasis, depression, and hysteria, compared to the general population and compared to patients with chronic heart disease.GLA, GLB1, NAGA, AGA, RPL36A-HNRNPH2, GAL, NOS3, UGCG, TTR, GAA, IDS, ELF3, TNF, ICAM1, UMOD, ABCB1, LAMP2, F5, VEGFA, TGFB1, NOS2, COL4A1, NOTCH3, TBC1D9, TTK, VCAM1, SOX2, TNNT1, SI, SMPD1, TLR4, SOD2, PRDX2, HNF1B, VDR, CDK5R1, VWF, UGT1A1, MIR451A, MIR423, MIR126, MPEG1, C16orf82, ATAD1, ACD, INF2, IGAN1, CHDH, PRKAG2, NPHS2, SMUG1, DAAM2, MCF2L, MLC1, SYNPO, RAPGEF5, ADIPOQ, MGAM, RPS27, CTSF, SELE, ACTB, PTGDS, DPP4, HNRNPA2B1, HNRNPA1, GUSB, SFN, G6PD, G6PC, FOS, F10, EPHA3, CTH, CTSA, CRP, SCARB2, CD80, BTK, ARSB, ARSA, APOE, AMBP, AGT, HSPA1A, HSPA1B, HSPA2, IDUA, PPBP, PON1, PODXL, PECAM1, NOTCH1, NOS1, NAIP, MYO5B, MPO, MEFV, MANBA, LRP2, LGALS3, LCN2, LAMP1, IL10, IL6, IL4, IL1A, NT5E
-
Leukodystrophy
Wikipedia
There is, however, a higher prevalence of the Canavan disease in the Jewish population. 1 in 40 individuals of Ashkenazi Jewish descent are carriers of Canavan disease. [28] This estimates to roughly 2.5%. Additionally, due to an autosomal recessive inheritance patterns, there is no significant difference found between affected males and affected females for most types of leukodystrophy including, but not limited to, metachromatic leukodystrophy, Krabbe disease, Canavan disease, and Alexander disease. ... Journal of Psychiatry and Neuroscience . 28 (3): 240. PMC 161748 . ^ Liu, Y; Zou, L; Meng, Y; Zhang, Y; Shi, X; Ju, J; Yang, G; Hu, L; Chen, X (June 2014). ... "Alexander's disease: reassessment of a neonatal form". Child's Nervous System . 28 (12): 2029–2031. doi : 10.1007/s00381-012-1868-8 .CLCN2, POLR3B, POLR3A, LMNB1, GJC2, GALC, CSF1R, TREX1, TUBB4A, POLR1C, IBA57, SDHA, HSPD1, DEGS1, DAG1, SCO2, NDUFV1, SURF1, SLC16A2, DARS1, RARS1, NKX6-2, ISCA2, ACOX1, EIF2B5, AIMP1, BOLA3, ARSA, COL4A1, PYCR2, TMEM106B, ACER3, ISCA1, FAM126A, IFIH1, AIFM1, NADK2, RNASEH2A, NDUFAF6, NDUFS7, CTC1, PEX16, SPG11, NUBPL, SLC19A3, EIF2S2, NDUFAF3, EIF2B2, EIF2B4, AIMP2, ALMS1, TUFM, SNORD118, PET100, RNASEH2B, SAMHD1, PNPT1, FBXL4, NDUFA12, TMEM126B, NDUFAF2, FOXRED1, PLEKHG2, MTFMT, NDUFB11, RNF216, NDUFA11, LIPT1, HIKESHI, TIMMDC1, GLRX5, TACO1, RNASEH2C, NDUFA13, NDUFAF5, NDUFAF4, SLC17A5, NDUFAF1, ABAT, NDUFB3, NDUFS3, NDUFA2, NDUFV2, NDUFS8, NDUFS6, NDUFA4, NDUFS4, NDUFA6, NDUFB9, NDUFS2, NDUFA9, NDUFS1, NDUFA10, COX15, NDUFB8, PC, NDUFA1, PDHA1, ND3, ND2, PEX1, ND1, ECHS1, SP110, EDNRB, EIF2B1, PLP1, ABCD1, PSAP, EPRS1, ADAR, NDUFB10, AARS2, GFAP, FA2H, ASPA, SDHB, MLC1, ABCB6, LGALS14, SOX10, RNASET2, PTHLH, GBE1, PDXP, SLC2A4RG, HEPACAM, COX10, MOG, PRDX5, ARHGEF2, CSF2, SOX17, ERCC6, ACOT12, CDSN, PLB1, CPS1, EIF2AK4, MIR219A1, MIR19B1, ATP5F1E, TYMP, ATN1, SARDH, COA8, CSH2, CSH1, ERCC8, LMNB2, OLIG1, TSEN54, CSF1, AQP4, RIEG2, GJA1, PLA2G6, SIGMAR1, OLIG2, TMEM63A, GDF15, NOTCH3, ATRN, PLA2G1B, POLR1A, YWHAZ, WT1, VTN, TYR, HSP90B2P, SORD, SDS, FGF21, SLURP1, ITGA2B, GJB1, GPM6B, HSD17B4, HSPA9, DARS2, RMND1, KARS1, NFU1, KIF5A, MAG, MBP, MPV17, SLC27A6, DCPS, LINC01672
-
Meconium Aspiration Syndrome
Wikipedia
"Meconium Aspiration Syndrome: Historical Aspects" . Journal of Perinatology . 28 : S3–S7. doi : 10.1038/jp.2008.162 . ^ Poggi, SH; Ghidini, A (2009). ... "Neurodevelopmental Outcome of Infants with Meconium Aspiration Syndrome: Report of a Study and Literature Review" . Journal of Perinatology . 28 : S93–S101. doi : 10.1038/jp.2008.154 . ^ a b Mokra, D; Calkovska, A (2013). ... "Extracorporeal Membrane Oxygenation: Use in Meconium Aspiration Syndrome" . Journal of Perinatology . 28 : S79–S83. doi : 10.1038/jp.2008.152 .