-
Colony Collapse Disorder
Wikipedia
These losses were later attributed to a combination of factors, including adverse weather, intensive apiculture leading to inadequate forage, Acarine (tracheal) mites , and a new infection, the chronic bee paralysis virus, [25] but during the outbreak, the cause of this agricultural beekeeping problem was unknown. [ citation needed ] Reports show similar behavior in hives in the US in 1918 [26] and 1919. [27] Coined "mystery disease" by some, [28] it eventually became more widely known as "disappearing disease". [29] Oertel, in 1965, [30] reported that hives afflicted with disappearing disease in Louisiana had plenty of honey in the combs, although few or no bees were present, discrediting reports that attributed the disappearances to lack of food. [ citation needed ] From 1972 to 2006, dramatic reductions continued in the number of feral honey bees in the US [31] and a significant though somewhat gradual decline in the number of colonies maintained by beekeepers.
-
Organophosphate Poisoning
Wikipedia
This test has been shown to be just as effective as a regular laboratory test and because of this, the portable ChE field test is frequently used by people who work with pesticides on a daily basis. [28] Treatment [ edit ] Current antidotes for OP poisoning consist of a pretreatment with carbamates to protect AChE from inhibition by OP compounds and post-exposure treatments with anti-cholinergic drugs. ... Archived (PDF) from the original on 2012-09-25 . Retrieved 2013-03-28 . ^ "Setting Tolerances for Pesticide Residues in Foods" . Archived from the original on 2013-04-01 . Retrieved 2013-03-28 . ^ "Food Quality Protection Act (FQPA) of 1996" . Archived from the original on 2012-11-04 . Retrieved 2013-03-28 . ^ "Children Are at Greater Risks from Pesticide Exposure" . ... Archived from the original on 2013-04-14 . Retrieved 2013-03-28 . ^ a b c d Human Rights Council.
-
Vascular Dementia
Wikipedia
A very large study conducted in Netherlands in 2015 found that the one-year mortality was three to four times higher in patients after their first referral to a day clinic for dementia, when compared to the general population. [26] If the patient was hospitalized for dementia, the mortality was even higher than in patients hospitalized for cardiovascular disease . [26] Vascular dementia was found to have either comparable or worse survival rates when compared to Alzheimer's Disease; [27] [28] [29] another very large 2014 Swedish study found that the prognosis for VaD patients was worse for male and older patients. [30] Unlike Alzheimer's disease, which weakens the patient, causing them to succumb to bacterial infections like pneumonia , vascular dementia can be a direct cause of death due to the possibility of a fatal interruption in the brain's blood supply. [ citation needed ] Epidemiology [ edit ] Vascular dementia is the second-most-common form of dementia after Alzheimer's disease (AD) in older adults. [31] [32] The prevalence of the illness is 1.5% in Western countries and approximately 2.2% in Japan.SDHAF1, BDNF, MAPT, IGF1, AVP, EPO, SST, GSK3B, FLT1, EIF4E, FGFR1, APOE, MTOR, CDK5, MMP2, TP53, GPX1, RPS6, SYK, MTHFR, CNTNAP2, NOTCH3, RASGRF2, ACE, CASP3, VEGFA, PON1, CSF2, TNF, LAMC2, PSEN1, BCL2, IL6, LPA, MMP9, GABPA, NFE2L2, ESR1, IL1B, COASY, TGFB1, ADIPOQ, CETP, AGT, CHI3L1, CRP, PIK3CG, NUBP1, PIK3CD, PIK3CB, HSPA4, ADM, GSTO1, TNFSF14, BCHE, CST3, SERPINE1, SYP, EDN1, TP63, PHLDB2, IL1A, ACHE, PIK3CA, ESR2, VWF, VDR, TXNRD1, AIMP2, SYN1, STAT5B, SREBF2, TXN, RPS6KA3, STAT5A, CCL2, SLC6A4, SLC19A1, TIMP4, SNCA, SOD1, SPP1, ADAM17, SNAP25, NAT2, GATD3A, RETREG1, TNFRSF19, RPGRIP1, HAMP, TRPV4, MOAP1, SPAG16, RIN3, MCHR2, CHRFAM7A, FOXP2, PTCRA, MCIDAS, MIR126, MIR134, MIR146A, MIR210, MIR222, OCLN, AD17, CRLS1, ANGPT4, SUCLA2, SAR1B, WASF1, CLDN1, RGN, KL, GRAP2, KLK4, WASF2, RFC1, AHSA1, TXNIP, NES, WASF3, RPGRIP1L, SYNM, CD2AP, RNF19A, ADGRA2, POLDIP2, NOX1, SIGMAR1, NTRK2, REN, GPI, COX8A, CLDN7, CREB1, CRK, MAPK14, DLG4, EGF, EPHA4, F11, FOXO1, FN1, FYN, GAP43, GC, GCG, GDNF, GFAP, CLU, CHRNA7, CHIT1, APOC3, ACTB, ADORA2A, ADRB1, AGER, AGTR1, ANK1, APOB, APP, CHAT, ARSA, ATF3, C5AR1, CASP1, CAV1, CBS, CFL2, GFRA1, GPT, PTPN1, GRN, NOS3, NOTCH1, SERPINA3, OXTR, PDE9A, PLA2G2A, PLG, PLEK, PON2, PPARG, MAPK1, PRNP, MASP1, KLK6, PTCH1, PTH, PTGS2, NMB, MAP3K5, MECP2, HTR6, GRIN1, GRP, HFE, HGF, HES1, HSPA1A, HSPA1B, IAPP, LCN2, ICAM1, IL4, IL10, IL18, JAK2, KDR, KRT6A, GATD3B
-
Euthyroid Sick Syndrome
Wikipedia
Mason expected the concentrations of thyroid hormones to rise in situations of psychosocial stress. [25] Mason's postulate was later confirmed by numerous studies. [26] [27] [28] [29] [30] [31] [32] [ excessive citations ] See also [ edit ] Thyroid storm References [ edit ] ^ a b Hershman JM.
-
Chronic Condition
Wikipedia
In 2008, the Trust for America's Health produced a report that estimated investing $10 per person annually in community-based programs of proven effectiveness and promoting healthy lifestyle (increase in physical activity, healthier diet and preventing tobacco use) could save more than $16 billion annually within a period of just five years. [25] It is uncertain whether school-based policies on targeting risk factors on chronic diseases such as healthy eating policies, physical activity policies, and tobacco policies can improve student health behaviours or knowledge of staffs and students. [26] Encouraging those with chronic conditions to continue with their outpatient ( ambulatory ) medical care and attend scheduled medical appointments may help improve outcomes and reduce medical costs due to missed appointments. [27] Finding patient-centered alternatives to doctors or consultants scheduling medical appointments has been suggested as a means of improving the number of people with chronic conditions that miss medical appointments, however there is no strong evidence that these approaches make a difference. [27] Nursing [ edit ] Nursing can play an important role in assisting patients with chronic diseases achieve longevity and experience wellness. [28] Scholars point out that the current neoliberal era emphasizes self-care, in both affluent and low-income communities. [29] This self-care focus extends to the nursing of patients with chronic diseases, replacing a more holistic role for nursing with an emphasis on patients managing their own health conditions. ... "National health spending by medical condition, 1996-2005". Health Affairs (Project Hope) . 28 (2): w358–67. doi : 10.1377/hlthaff.28.2.w358 . ... "In chronic condition: experiences of patients with complex health care needs, in eight countries, 2008" . Health Affairs . 28 (1): w1-16. doi : 10.1377/hlthaff.28.1.w1 .
-
Amaurosis Fugax
Wikipedia
However, it is also not uncommon for these patients to have no other symptoms. [18] One comprehensive review found a two to nineteen percent incidence of amaurosis fugax among these patients. [19] Systemic lupus erythematosus [20] [21] Periarteritis nodosa [22] Eosinophilic vasculitis [23] Hyperviscosity syndrome [24] Polycythemia [25] Hypercoagulability [26] Protein C deficiency [27] Antiphospholipid antibodies [28] Anticardiolipin antibodies [29] Lupus anticoagulant [30] [31] Thrombocytosis [29] Subclavian steal syndrome Malignant hypertension can cause ischemia of the optic nerve head leading to transient monocular visual loss. [32] Drug abuse-related intravascular emboli [8] Iatrogenic : Amaurosis fugax can present as a complication following carotid endarterectomy , carotid angiography , cardiac catheterization , and cardiac bypass . [29] Ocular origin [ edit ] Ocular causes include: Iritis [33] Keratitis [24] Blepharitis [24] Optic disc drusen [29] Posterior vitreous detachment [24] Closed-angle glaucoma [34] Transient elevation of intraocular pressure [8] [33] Intraocular hemorrhage [8] Coloboma [29] Myopia [29] Orbital hemangioma [35] Orbital osteoma [36] Keratoconjunctivitis sicca [29] testing Neurologic origin [ edit ] Neurological causes include: Optic neuritis [8] Compressive optic neuropathies [8] [29] Papilledema : "The underlying mechanism for visual obscurations in all of these patients appear to be transient ischemia of the optic nerve head consequent to increased tissue pressure.JAK2, ADRA2B, CNTN2, TRNQ, TRNS1, TRNS2, TRNW, NOTCH3, PIK3CA, PMS1, PMS2, RPS20, MLX, TRNH, TGFBR2, TP53, SH2B3, FAN1, PTPN22, MLH3, ADA2, TET2, SEMA4A, TRNL1, TRNF, BMPR1A, ND6, CALR, CTNND2, MSH6, HLA-B, IL12B, KRAS, EPCAM, MLH1, MPL, MSH2, COX1, COX2, COX3, ND1, ND4, ND5, SAMD12, MMP1, SPP1, GIP, F5, F2
-
Compulsive Hoarding
Wikipedia
., co-morbid OCD), and OCD-based hoarding. [26] Given the aforementioned distinction, it was proposed to increase coverage of compulsive hoarding in the forthcoming [ needs update ] Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-V), both by creating a distinct category for compulsive hoarding, provisionally named, Hoarding Disorder [27] (either in the main manual under " obsessive-compulsive spectrum disorders" or in the appendix), and by including hoarding as a potential symptom of OCD. [28] Book hoarding [ edit ] Main article: Bibliomania Bibliomania is a disorder involving the collecting or hoarding of books to the point where social relations or health are damaged. ... "Comorbidity in hoarding disorder" . Depression and Anxiety . 28 (10): 876–884. doi : 10.1002/da.20861 .
-
Bdsm
Wikipedia
Physically, BDSM is commonly mistaken as being "all about pain". [25] Freud was confounded by the complexity and counterintuitiveness of practitioners' doing things that are self-destructive and painful. [26] Most often, though, BDSM practitioners are primarily concerned with power, humiliation, and pleasure. [25] The aspects of D/s and B/D may not include physical suffering at all, but include the sensations experienced by different emotions of the mind. [25] Of the three categories of BDSM, only sadomasochism specifically requires pain, but this is typically a means to an end, as a vehicle for feelings of humiliation, dominance, etc. [ citation needed ] In psychology, this aspect becomes a deviant behavior once the act of inflicting or experiencing pain becomes a substitute for or the main source of sexual pleasure. [27] In its most extreme, the preoccupation on this kind of pleasure can lead participants to view humans as insensate means of sexual gratification. [28] Dominance and submission of power are an entirely different experience, and are not always psychologically associated with physical pain. ... According to yet another survey of 317,000 people in 41 countries, about 20% of the surveyed have at least used masks, blindfolds or other bondage utilities once, and 5% explicitly connected themselves with BDSM. [97] In 2004, 19% mentioned spanking as one of their practices and 22% confirmed the use of blindfolds or handcuffs. [97] A 1985 study found 52 out of 182 female respondents (28%) were involved in sadomasochistic activities. [98] Recent surveys A 2009 study on two separate samples of male undergraduate students in Canada found that 62 to 65%, depending on the sample, had entertained sadistic fantasies, and 22 to 39% engaged in sadistic behaviors during sex.
-
False Memory
Wikipedia
Regardless of the effect being true or false, the respondent is attempting to conform to the supplied information, because they assume it to be true. [28] Loftus' meta-analysis on language manipulation studies suggested the misinformation effect taking hold on the recall process and products of the human memory. ... Clinical Psychology: Science and Practice . 11 : 3–28. doi : 10.1093/clipsy.bph055 . ^ Knafo, D. (2009). ... Discover Magazine . Retrieved 12 December 2019 . ^ 28 December 1994, on the cable channel TNT ; the marathon featured movies including Sinbad and the Eye of the Tiger (1977). ^ Evon, Dan (28 December 2016).
-
Sepsis
Wikipedia
If other sources of infection are suspected, cultures of these sources, such as urine, cerebrospinal fluid, wounds, or respiratory secretions, also should be obtained, as long as this does not delay the use of antibiotics. [10] Within six hours, if blood pressure remains low despite initial fluid resuscitation of 30 ml/kg, or if initial lactate is ≥ four mmol/l (36 mg/dl), central venous pressure and central venous oxygen saturation should be measured. [10] Lactate should be re-measured if the initial lactate was elevated. [10] Evidence for point of care lactate measurement over usual methods of measurement, however, is poor. [28] Within twelve hours, it is essential to diagnose or exclude any source of infection that would require emergent source control, such as a necrotizing soft tissue infection, an infection causing inflammation of the abdominal cavity lining , an infection of the bile duct , or an intestinal infarction. [10] A pierced internal organ (free air on an abdominal x-ray or CT scan), an abnormal chest x-ray consistent with pneumonia (with focal opacification), or petechiae , purpura , or purpura fulminans may indicate the presence of an infection. ... Those without multiple organ system failure or who require only one inotropic agent mortality is low. [85] Other [ edit ] Treating fever in sepsis, including people in septic shock, has not been associated with any improvement in mortality over a period of 28 days. [86] Treatment of fever still occurs for other reasons. [87] [88] A 2012 Cochrane review concluded that N-acetylcysteine does not reduce mortality in those with SIRS or sepsis and may even be harmful. [89] Recombinant activated protein C ( drotrecogin alpha ) was originally introduced for severe sepsis (as identified by a high APACHE II score), where it was thought to confer a survival benefit. [78] However, subsequent studies showed that it increased adverse events—bleeding risk in particular—and did not decrease mortality. [90] It was removed from sale in 2011. [90] Another medication known as eritoran also has not shown benefit. [91] In those with high blood sugar levels, insulin to bring it down to 7.8–10 mmol/L (140–180 mg/dL) is recommended with lower levels potentially worsening outcomes. [92] Glucose levels taken from capillary blood should be interpreted with care because such measurements may not be accurate. ... During the same time frame, the in-hospital case fatality rate was reduced from 28% to 18%. However, according to the nationwide inpatient sample from the United States, the incidence of severe sepsis increased from 200 per 10,000 population in 2003 to 300 cases in 2007 for population aged more than 18 years.NOS2, TLR4, ANGPT1, CASP3, CSF3, HMGB1, IFNG, ADM, TNF, PROC, IL10, LTA, LBP, MAPK1, TGFB1, NOS3, TGFBI, MMP9, CXCR2, MIF, ADORA2B, MAPK3, IFNA2, ADORA2A, BCL2, BTBD8, LCN2, PCSK9, SIRT1, MBL2, TEK, FCGR1A, MPO, THBD, CALCA, TLR2, IL1RN, COTL1, HMOX1, CD14, IL33, IL1A, IL1B, HIF1A, UROD, ADAMTS13, IL6, CASP1, CXCL8, CALML3, MYD88, IL17A, IL18, F3, NLRP3, CCL2, NM, PIK3CD, MIR146A, PDCD1, ELOVL4, SERPINE1, PIK3CG, ANGPT2, ALB, TREM1, TLR9, XIAP, CSRP3, NFE2L2, AGER, CRP, MIR21, STAT3, PTX3, CD274, PIK3CA, C5AR1, GPT, ITGAM, ACTB, PIK3CB, VWF, GSDMD, ICAM1, CIRBP, MIR155, XRN1, GABPA, ACE, NOD2, MAPK8, SEPTIN1, TLR1, CFH, SDC1, MIR223, F5, CD86, RETN, AQP5, IL4, PRKAA1, COPD, PRKAA2, ROS1, PPARG, VEGFA, AVP, AZU1, MALAT1, P2RX7, S100A8, HP, F2R, CSF2, HLA-DRA, WNT5A, HPSE, F2, ENTPD1, S100A9, ITGB2, HRG, BCHE, SMPD1, IL17D, PROCR, PRKAB1, VPS51, IL7, IFNA1, TNFRSF1A, PTGS2, SELE, CAV1, NFKB1, IFNA13, CD163, SPP1, MYDGF, DECR1, MIR150, PLA2G2A, LY96, SFTPD, REG1A, RELA, MYLK, MFAP1, HAMP, MMP8, IFNB1, TXN, IL2RA, NR3C1, VCAM1, IRAK1, ISG20, MTOR, IRAK3, TLR3, RIPK3, IL27, PADI4, MFGE8, NEAT1, CTLA4, UCP2, AKT1, APRT, ADIPOQ, KL, ALK, GDF15, C5, MIR23B, CASP4, CD69, CFB, AIMP2, AHSG, EGF, MIR126, LEP, MIR122, POLDIP2, GORASP1, MEFV, PDE4B, AMBP, RNF19A, PC, THPO, TRPM7, CAMP, CCR2, EDN1, GCG, BDNF, EOS, CXCR4, GAS6, SYT1, SIRT3, FCGR3A, APOA1, SIRT2, WNK1, CEBPB, CXCL1, MBL3P, ADAM10, HSPA4, ACHE, CYBB, AHSA1, CRK, TIRAP, MAPK14, SELP, IL22, PON1, SAA1, IL2, SIRT6, S100B, GRAP2, GSN, MIR143, JUN, JUNB, SLC52A2, JUND, TNFRSF6B, BAX, NAMPT, JAK2, HOXD13, APLN, PDCD1LG2, TNNI3, LGALS3, CAT, ABCC11, CARS1, ACCS, IGF1, NR1I2, CD19, BPI, CD80, SPHK1, IDO1, MINDY4, HSPD1, HAVCR2, TM7SF2, KAT8, CST3, TFPI, PINK1, PPIG, PLA2G1B, PLAU, PLD1, PLG, SLC2A1, TLR7, GP6, HDAC6, SETMAR, PTEN, PTH, CXCL12, ADAR, FOXP3, MTCO2P12, LINC02605, C5AR2, RPS6KB1, SAA2, S100A1, SMOX, SOAT1, ATG16L1, CD177, ATP5F1A, ATP2A2, SERPINC1, HOTAIR, NR3C2, MMP1, TAT, SEMA6A, TAC1, COX2, PPP6R3, ABCC8, PLA2G15, GZMB, NDUFA2, FAS, ACSS2, NOS1, PANX1, LOC100505909, AQP1, PPP6R2, ELANE, LAT2, DPYD, DPYS, VDR, MIR26B, MIR145, CETP, CHI3L1, VIP, MIR142, FOSB, S1PR3, GJA1, EZH2, FOXM1, RTL1, PPM1D, PPARGC1A, PWAR1, MIR25, BTLA, FOS, BECN1, CTH, FASN, SCGB1A1, CXCL2, FCGR3B, P2RY1, ARG1, MEG3, TNFRSF11B, ORM1, P2RX4, OCLN, MMP13, APOL1, SOD2, FCGR2B, ARSA, SNCA, NUP62, ACE2, CASP12, PRKN, FER, EWSR1, FCMR, SSRP1, F2RL1, APOM, CNR2, MYC, AQP3, MSC, SOCS1, SLBP, C20orf181, FST, MIF-AS1, DNM1L, HCAR2, SYP, ERN1, LINC01672, NOTCH1, FCAR, COX8A, FCGR2A, GTF2H1, TACR1, MRC1, QPCT, RPL17-C18orf32, GTS, PENK, PF4, CRH, PTMA, PTMAP4, RN7SL263P, IL37, MIR15A, MIR17, HEBP1, MIR182, RAC1, MOK, RAG1, KHDRBS1, PRPH2, SPATA2, RELB, CCL1, KEAP1, SETD2, RPL17, CTSB, CTNND1, PLA2G10, SLC25A10, CLOCK, IRAK4, SELENOP, DNMT1, POMC, PFKFB3, ALPL, ALDH2, MIR125A, PKM, EDNRA, EMSLR, PLAT, PLAUR, MASP2, IL23A, DUSP1, DCTN4, HAVCR1, TNFRSF12A, PPARA, PPARD, TSC22D3, CLEC1B, PRKCA, PRKCB, GRK2, MFN2, EIF2AK2, TRPV4, ADAM17, IL13, TLR5, IL15, GFAP, SQSTM1, TRPV1, CXCL10, NCF1, TMSB4X, HSP90AA1, GC, HSPA1B, MAP1LC3B, TICAM1, ITGAL, PLA2G6, HSPA1A, ITGB1, BRCA1, KLF2, TRPM2, GAPDH, MIR608, CD38, TNC, TP53, SIRT5, GPR174, CD22, TNFSF13, CHRFAM7A, STAM2, TSLP, HSH2D, CCK, CBS, VIM, CASP9, IL3, MIR193B, TRIM69, IL5, GLP1R, GJA5, VLDLR, GIP, CXCR1, GH1, C3AR1, G6PD, CD5L, FOXO1, SLC39A14, CHRNA4, TAGAP, TNFRSF13B, SMAD2, MB, FN1, KDM6B, TGFBR2, LAG3, FPR2, CD74, CBLL2, PARK7, BAAT, CHRNA7, PAGR1, CD59, HDLBP, B2M, TIMP1, NR4A1, TFAM, LTF, DDAH2, CD40LG, HSPA12B, KNG1, STAB1, MUL1, SOCS3, TST, MBTPS1, LDLR, BBC3, NLRP1, OLFM4, RAMP2, LILRB2, TNFSF13B, SARM1, P2RX2, PDCD4, NOX1, AAK1, TXNIP, SIGLEC9, NBEAL2, TLK1, ANP32B, RAMP1, IL36A, NR1I3, CAMKK2, PDPN, TLR6, ABRAXAS2, RRS1, FARP2, CILK1, CTCF, NAAA, SULF1, SIGMAR1, IL17C, TRIM22, CXCR6, CD2AP, NR1H4, EBI3, MICU1, IL17RA, CADM1, STK38, SLC9A6, GPR182, IL24, BCL2L10, SMC4, SH3BP4, LILRB3, UBAC1, NR1H3, CORO1A, LPAR3, EDIL3, ESM1, BCAP31, LRPPRC, DDAH1, DLC1, NOD1, RASGRP1, GCA, TNFAIP8, PUF60, SYNCRIP, HDAC5, RIDA, ACAD8, FOXD3, FGF21, TRAF3IP2, FBXO3, PRG4, ATG7, DAPK2, HCST, SENP6, SETBP1, KAT5, SPATS2L, RALBP1, PRDX3, MGLL, SUMF2, SDS, CALCRL, PSME3, ARIH1, MPHOSPH6, A2M, SGSM3, MIR19A, IL31, MIR130B, MIR140, MIR141, MIR148A, MIR152, MIR181A2, MIR184, MIR187, MIR191, MIR199B, MIR200A, MIR34A, MIR210, MIR211, MIR216A, MIR23A, MIR27A, MIR27B, MIR29A, MIR29B1, MIR29B2, MIR30A, MIR30E, ASPG, NDUFS7, PWAR4, IRGM, BPIFA2, CTCFL, RSS, OR2AG1, PTGR2, IL34, CD300LF, IL23R, PLB1, RMDN2, MARCHF10, OXER1, SGMS2, MCEMP1, NRK, GPRC6A, IPMK, MRGPRX1, OR10A4, CLEC4D, STING1, TREML1, COL6A4P1, MIR31, MIR7-1, MAGEC3, PGR-AS1, CDKN2B-AS1, ZGLP1, MIR300, MIR874, MIR297, CD24, MIR1260A, MIR3165, TMX2-CTNND1, P2RX5-TAX1BP3, MIR4772, MIR6835, MIR7-2, MIR7110, THRIL, CBSL, LOC102725035, ERICD, NKILA, LARP1BP2, RNU6-392P, LOC107987462, H3P24, H3P47, TLX1NB, ACOD1, SFTPA2, HULC, MIR7-3, MIR98, VN1R17P, ZFAS1, GPR166P, MIR335, MIR346, MIR370, LNCR1, MIR375, H4C15, HNP1, MIR146B, MIR494, MIR495, MIR499A, MIR505, MIR483, UCA1, SFTPA1, PPAN-P2RY11, MIR590, DEFA1B, FUNDC1, GPR151, IGHV1-69, IL26, EGLN1, UGT1A1, TRPM4, DYM, TENT5C, TUG1, ANO1, RMDN3, SPTLC3, SYBU, IFT122, ADCY10, RPTOR, SELENOS, HDAC8, PBK, TRPV5, PPAN, DUSP22, FAM20C, ACKR3, AGTRAP, PNPLA2, ZNFX1, TOLLIP, MIEF1, CYTL1, SUFU, DLL1, HIPK2, SENP1, IL19, ADGRE2, TAX1BP3, NOX4, AK3, SOST, TMED5, ASCC1, GLRX2, RMDN1, SDF4, ZNF580, PLEKHO1, HSPA14, CRBN, TLR8, SLC25A37, PLAC8, ISYNA1, GDE1, MTA3, CFAP97, TNFAIP8L1, C1QTNF1, SYVN1, IL1F10, MFSD2A, SYS1, MYLK3, TIMD4, G6PC3, NAPRT, PPP1R14A, PRRT2, CMTM7, C1QTNF3, NLRC4, FBXO32, TNFRSF13C, IL22RA2, SLC18B1, MRGPRX3, MRGPRX4, SLC16A10, LRRK2, H4-16, MTFMT, CD300LB, TCHP, CRISPLD2, JAM3, VOPP1, IL21, LGR6, GAS5, SLC39A8, TINAGL1, ZDHHC6, CLEC7A, CDK15, MARCKSL1, AACS, COLEC11, MAPKAP1, GIT2, EFHD2, MMEL1, CORO7, NLRX1, EHMT1, ADM2, TNIP3, ZC3H12A, WDR26, COL18A1, LILRA6, SLC7A2, HDAC4, SMAD3, GAST, FUT4, ACKR1, GALNS, GATA3, GBP2, GDNF, GEM, GJA4, GLUL, GPR15, GPR18, GPR42, GRK5, GPX3, GPX4, GRIN1, GRIN2A, GRIN2B, GRP, GRPR, GSK3B, GSR, GZMK, HABP2, FTH1, FSHMD1A, FPR1, FABP4, ENG, EPHB2, EPO, ERBB4, ESRRB, ETS2, EXTL3, F8, F9, F10, F13A1, FABP3, FOSL2, ACSL1, ACSL4, PTK2B, FCN1, FCN2, FGFR1, FH, FOXO3, FLI1, FLII, FLNA, HCRT, HDAC2, NRG1, KIR3DS1, ITGA3, ITGAX, ITGB3, ITGB4, ITGB5, ITPR3, KCNJ5, KCNJ8, KCNMA1, KDR, KIR3DL1, KLKB1, IRF6, KLRC1, KPNA1, TNPO1, KRT18, LAMP1, LAMC2, LIPE, LPA, LRPAP1, CYP4F3, LY6E, ITGA2B, IRF3, HLA-B, IFNAR1, HMGCR, HMGA1, TLX2, HPRT1, HSD11B2, HSPA5, HSPB1, HSPE1, IAPP, IFI27, IFN1@, IFNAR2, IRF1, IGFBP6, IGFBP7, CCN1, IGHG3, IL3RA, IL6ST, IL7R, IL9, IL10RA, IL12RB1, IREB2, MARK2, ELAVL2, EIF4EBP1, BCL6, FASLG, AQP2, AQP4, AQP9, ARRB2, ASS1, ATF3, ATP1A3, ATP2A1, BCKDHA, CCND1, PRDM1, AQP8, BLVRA, BRS3, BTK, BUB1, BYSL, TSPO, SERPING1, CALR, CASP5, CASP8, CD3D, APOH, APOC1, CD27, AFP, ABO, ACAT1, ACR, ACP3, ACTA1, PARP1, ADRA1A, ADRA2B, ADRB2, ADRB3, AFA, JAG1, APOB, AGT, AGTR1, AGTR2, AIF1, AKR1B1, ALOX12, ALOX15, AMH, AMPD1, AOAH, APC, CD247, CD28, EPHA2, NQO1, CCN2, CTSG, CTSZ, CX3CR1, CYP1A2, CYP2C8, ECI1, DCR, DDT, DEFA1, DHCR24, DIO1, CSRP2, DLD, DNASE1, DPEP1, SLC26A3, RCAN1, DUSP3, DUSP6, EDA, S1PR1, EDNRB, EGR1, CSTB, CSE1L, CD34, ACKR2, CD40, CD44, CDH5, CDK9, CDKN1C, CETN1, CGA, CHIT1, CHRM3, CISH, CCR5, CNR1, CS, COMP, MAP3K8, COX5B, CP, CPB1, CPN1, CR1, CREB1, CREBBP, CRIP2, CRMP1, LYZ, MAS1, BCAR1, MAT1A, TRAF6, HSP90B2P, TSC2, TYK2, UBA1, UCHL1, UCP1, UPP1, UQCRFS1, UTRN, UVRAG, VTN, XBP1, XPO1, YY1, YWHAZ, ZAP70, ZFP36, DNALI1, IL1R2, SCG2, NELFE, PLA2G7, TFPI2, GLRA3, HSP90B1, TOP2A, TOP1, STXBP3, SLC22A1, SLPI, SMS, SOX10, SPN, SPOCK1, SST, SSTR1, SSTR4, ST2, SULT1E1, TAPBP, CLDN5, TCF21, TRBV20OR9-2, PRDX2, TEAD4, TERF2, TERT, TFDP1, TFRC, TH, THBS1, TIMP2, HMGA2, TAM, H4C9, XPR1, CFLAR, PROM1, ARHGEF7, MBD2, WASL, F2RL3, MAP3K14, NMI, P2RX6, PCSK7, LPAR2, IL32, IL18RAP, GPR55, S1PR2, KLF4, TMPRSS11D, NTN1, GSTO1, PICK1, ATG5, TBPL1, GOSR1, CIR1, NRP1, TNFRSF11A, FZD4, ULK1, H4C1, H4C4, H4C6, H4C12, H4C11, H4C3, H4C8, H4C2, H4C5, H4C13, H4C14, ST11, RAB11A, CAMK1, LGR5, MAPKAPK5, DENR, EIF3B, MARCO, TNK1, RIPK1, CRADD, TNFSF10, ADAM15, SLC16A2, SLC15A1, ABCF1, P2RY11, NPPB, NPPC, NPR3, YBX1, NT5E, NTS, NR4A2, P2RX1, P2RX3, P2RX5, P2RY2, PAEP, NID1, PRDX1, PAWR, PBX3, PCSK1, CDK16, PDE4D, PDR, SERPINF1, ABCB1, SERPINA1, PIN1, NPHS1, NHS, PKD1, MPZ, MC1R, MCL1, CD46, MDK, MDM2, ATXN3, MMP2, MMP3, MMP14, MMP16, MPST, MSMB, NFE2, MSN, MST1, MT2A, COX1, MTHFR, MTTP, MUC2, NCAM1, NDUFS3, NEDD4, NFATC3, PITX3, PLA2G4A, SLC5A4, S100A12, OPN1LW, REL, REN, RFC1, RNASE1, RPL4, RPL9, RPS6KA3, RRBP1, RYR1, RYR2, ATXN7, KDM5A, SCN1A, SCN10A, CCL4, SEA, SELL, SFRP1, SFTPB, ITSN1, SLC2A4, SLC5A1, SLC5A2, RBP4, RAB1A, PLCG1, PRKACG, PLTP, PLXNB1, PNOC, POR, POU2F1, PPIA, PPP1R1A, PTPA, PRF1, PRH1, PRH2, PRKCD, PVT1, MAP2K3, PSPN, PROS1, MASP1, PSAP, PSMD10, PSPH, PTPN1, PTPN11, PTPRC, PVR, H3P40
-
Atherosclerosis
Wikipedia
This somewhat offsets the narrowing caused by the growth of the plaque, but it causes the wall to stiffen and become less compliant to stretching with each heart beat. [25] Modifiable [ edit ] Bacterial infections [26] Diabetes [27] Dyslipidemia [27] Tobacco smoking [27] Trans fat [27] Abdominal obesity [27] Western pattern diet [27] Insulin resistance [27] Hypertension [27] HIV/AIDS [28] Nonmodifiable [ edit ] Advanced age [27] [29] Male [27] Genetic abnormalities [27] Family history [27] Coronary anatomy and branch pattern [30] Lesser or uncertain [ edit ] South Asian descent [31] [32] Thrombophilia [33] [34] [35] Saturated fat [27] [36] Excessive carbohydrates [27] [37] Elevated triglycerides [27] Systemic inflammation [38] Hyperinsulinemia [39] Sleep deprivation [40] Air pollution [41] [42] Sedentary lifestyle [27] Arsenic poisoning [43] Alcohol [27] Chronic stress [27] Hypothyroidism [44] Periodontal disease [45] Dietary [ edit ] The relation between dietary fat and atherosclerosis is controversial.LDLR, AGT, AGER, HP, SIRT1, PPARG, CYP27A1, CYBA, CRP, CETP, ADIPOQ, VEGFA, CAV1, APOE, APOA4, ALOX15, ALOX5, ESR1, STAT3, VCAM1, TLR4, ICAM1, IFNG, IL6, LPA, CCL2, MMP1, PTGS2, NOS3, SERPINE1, PON1, TNF, PLA2G2A, ABCA1, GSTM1, ALOX5AP, SOCS3, PLAT, CNR2, AHR, ABCG5, SOD1, NR1H4, NOS2, PARP1, IGF2, LTA4H, CCL4, SOD2, HRH1, TRIM28, BCL3, CELA2A, NECTIN2, HSPA1B, CCL3, TRPV1, FASLG, NPC1, CD40LG, IL18, CD40, CD36, MMP2, SELP, TTPA, HMGB1, CLU, F2, CASP3, CASR, FAS, LCAT, LMNA, LDLRAP1, ENPP1, ENG, FOS, WRN, SLC17A4, MIR21, HSPA14, GCG, CRK, FGF21, BANF1, DPP4, ANGPTL4, PLA2G7, KLF2, GGCX, COL3A1, TP53, GABPA, GLP1R, TM7SF2, TLR2, NR1I2, EDN1, THBS1, TGFBR3, TGFB1, CCR5, POLDIP2, HGD, EGR1, BSCL2, FGF23, MAPK14, DECR1, F3, HIF1A, ACE, AIMP2, ANGPTL3, VWF, FABP4, MIR33A, NOX4, IL37, PLA2G10, CX3CR1, UCP2, FOXP3, DNMT1, TXN, FN1, ALMS1, TNFSF4, MIR155, CST3, ELN, CXCR4, UTS2, HMOX1, NOTCH3, PRKAA1, PRKAA2, PRKAB1, MAPK1, TNFRSF11B, OLR1, NPY, REN, IL33, NFE2L2, PTX3, MOK, NLRP3, ACE2, AHSA1, ANTXR1, PAPPA, PPIA, ANGPTL6, PPARA, PON2, PLTP, LPAL2, PLG, PECAM1, PIK3CA, PIK3CB, PIK3CD, PIK3CG, EHMT1, PLA2G1B, MTHFR, CXCL16, TET2, CAVIN1, PCSK9, CHDH, SPP1, IL17A, IL10, RNF19A, AGPAT2, MPO, SREBF2, IL1B, IL1A, PINX1, IGF1, HSPD1, SLC5A2, APOM, RETN, SELE, LEP, LGALS3, CXCL12, CX3CL1, LPL, LRP1, MBL2, SAA1, SORT1, MEFV, MMP3, MMP9, MMP12, CXCL8, MIR126, KLF4, XPR1, APOH, AKT1, CD44, LIPG, AGXT, ALB, CCR2, ABCC6, SAMD4A, ABCG1, AHSG, GRAP2, ANGPT1, NR1H3, VPS51, CAD, ATM, SMUG1, CDKN2A, CDKN2B-AS1, ZGLP1, CHI3L1, APOA2, APOB, ANGPT2, SCARB1, APOA1, CD14, AGTR1, KL, CFDP1, APOC3, ADAMTS7, MALAT1, GPR162, INSR, APOL1, MAPK3, TNC, BSG, SLC2A1, CTSS, TNFRSF9, ANPEP, SOST, MTCO2P12, HAMP, EDNRA, ALDH2, MYD88, MIR221, GJA4, APOA5, ESR2, COX2, MIR145, CXCR6, IL23A, MTOR, WNT5A, USF1, MIR146A, CD68, PON3, YWHAZ, ADAM17, ACTB, CASP1, MIR223, PADI4, HMGCR, CIMT, PPARD, LCN2, IL4, LTA, CCL5, ACAT1, S100A12, ITGAX, CCN2, NR3C2, PLB1, ABCB6, ACKR3, BECN1, MFAP1, ROS1, BMS1, ACACA, APRT, AKR1B1, PTPN22, GDF15, FOXO1, RENBP, ADRA2B, HSPB3, FGF2, FCGR3A, CD34, NAMPT, HSPB2, NR4A1, MIR27A, TSPO, BCL2, HSPB1, COPD, KDR, SGMS2, SOAT1, TCF7L2, STAT1, SFTPD, F11R, INS, SSTR4, BGN, MMP13, ADRA1A, PGR-AS1, IFNA1, BGLAP, FOXO3, IFNA13, BRS3, RARRES2, LGMN, HSPA4, TRAF6, CAT, P2RY2, SELPLG, TFPI, HGF, HDAC9, LIPC, TIMP1, IL2RA, TREM1, GPR42, SAA2, JAK2, CAMP, ITGB2, PTH, FTO, CLOCK, HSD11B1, GSTT1, GAS5, VCAN, SQSTM1, F2RL1, PDE4D, LPAR2, HPSE, APLN, PLPP3, TFEB, TFPI2, TNFSF10, CSF2, KNG1, MIRLET7G, EGFR, ALOX15B, GNB3, CTNNB1, TNFRSF12A, TNFRSF1A, AOC3, MIR122, MMRN1, SIRT3, PTEN, MAPK8, GH1, NOTCH1, ABO, PDGFD, CCL11, MYDGF, CXCL1, GPX1, PLA2G6, MIR29A, HSPG2, NTN1, C1QTNF9, P2RX7, TERT, IGF1R, MIR34A, TUG1, SREBF1, PGF, THBD, ECE1, NT5E, PF4, CYBB, ISG20, IL32, CDKN2B, GRN, ADIPOR1, TOR2A, CD1D, S100A8, EZH2, SCD, SLC33A1, APP, MMP8, FCGR3B, DHX40, MIF, FMO3, MGP, VLDLR, EPRS1, F2R, ARG1, ABCG8, CCR7, MIR217, CHIT1, TNFSF13B, IL9, LPCAT3, IL6ST, CYP2C9, MIR30C1, BMP2, IL13, CES1, NFKB1, IDO1, MSR1, MYOCD, PDZK1, CHRM3, LTB4R, MIR30C2, IL1RN, ADIPOR2, VKORC1, PKM, AIF1, SERPINF1, MTTP, CCN1, RBP4, NOD2, CD163, RNR2, SGSM3, NOS1, MMP14, COMP, LOX, RYR3, PTGIR, KLF14, S100A9, IL23R, NUP62, ACLY, MAP3K5, PLIN2, PLAU, SOCS1, LRP6, IRS2, ARNTL, AQP1, CTH, IRAK1, PTGS1, OR10A4, TRIB3, IL25, IL27, PHACTR1, SMPD1, SLC9A1, MFGE8, ANGPTL8, DKK1, CP, APLNR, TNFSF12, SHBG, F10, TIMP3, CD59, KHDRBS1, FLNA, MIR17, MIR590, DDR1, MAP4K4, CNBP, EPHB2, FLT1, MIR24-1, HFE, PPARGC1A, UCP1, CFH, IL17D, GAS6, GHRL, ENTPD1, FURIN, MIR210, IL19, CD47, GCH1, TNFRSF4, S1PR1, GJA1, VTN, ADAMTS4, DNER, TLR7, EPHX2, GHSR, FPR2, GTF2H1, ADM, MIR19B1, ELANE, TCF21, SIRT6, VIM, DCTN4, ELAVL2, FCGR2A, P4HB, S100B, MAZ, MAS1, YAP1, MAOA, PNPLA2, MIR216A, MIR144, GIP, LIPA, CCL22, EPHA2, PTGDS, ANXA5, CRISP2, ELK1, AOC2, MIR377, ANGPTL2, GC, NISCH, MIR143, FSHMD1A, XDH, SYK, AR, ADAM10, VDR, MT2A, F5, COX1, PTPN1, APOBEC1, FBN1, MIR19A, PTK2B, ACSL1, TRPV4, MYC, FABP5, MIR20A, ADAMTS13, MYLK, MIR33B, APOA1-AS, CREG1, CAPN10, MRC1, MBTPS1, CBS, MIR212, APCS, LEPR, NCEH1, ESAM, ROCK1, XBP1, ADAM15, SEMA6A, MERTK, JCAD, GAL3ST1, EVPL, PIN1, FGF13, CD5L, BMP4, LINC01672, NEAT1, HBEGF, IKBKB, TNFSF12-TNFSF13, DUSP1, CA2, NOS1AP, SERPIND1, PROS1, PSRC1, SLC52A1, IL15, MIAT, CST12P, CYP2B6, SEMA7A, HABP2, SLC35A1, TIE1, LRP8, MIR98, CASZ1, HPGDS, OGA, BTF3P11, SETD2, HSPA5, TAGLN, MIR497, HSP90AA1, KLF5, IL1F10, NR4A3, STAT4, DYM, ID3, MAP2K1, DCN, RMC1, PCLAF, HLA-C, HCAR2, IRF1, CBSL, MEG3, NPPC, TP53COR1, IRF6, MIR222, B2M, CTSD, NOX1, TNFSF11, KIT, ADAM8, GPI, ANXA1, PRDX1, MIR10A, SFRP5, SCAP, EGF, TNFSF15, GPBAR1, STC2, NPPB, TLR5, SP6, BATF3, CTSK, ITGB3, GSK3B, ADD1, CXADR, ITGA2B, SORL1, KIF6, SLC25A20, PLIN1, TRIM13, COPS5, ZMPSTE24, MAPK7, PRTN3, NOX5, PRG4, PPBP, PDIA2, GORASP1, GDF11, PLXNA2, ELOVL6, WNK1, NOD1, PROC, PLAUR, PRKAR1A, CLEC7A, RIPK3, PTAFR, LDB2, NIF3L1, AIM2, IL27RA, PADI1, ZFP36, XIST, ROCK2, ADAMTS3, NOXA1, VCL, IL20, MBL3P, CCL4L2, TUFM, PLA2G4A, TMED7, TRPC3, IRAK4, NCOR1, ZC3HC1, TNFRSF1B, NFAT5, TLR3, NPC1L1, RGCC, RAG1, TAM, TBX18, POU2F3, ABCA12, HDAC3, PLA2G15, RIPK2, HSPB8, TNFSF9, SLC17A5, TNFSF14, GREM1, PDE5A, FOXP1, DKK3, PALD1, RGS5, SOAT2, INTU, AIMP1, LOH19CR1, NAAA, THBS4, TGM2, NR2F2, TFAP2A, NR1I3, KLRK1, ZHX2, HDAC5, CCL21, PDCD6IP, CCL19, CCL18, PLAAT1, AS3MT, AHRR, S100A1, RXRA, RPS19, RORA, RNF213, NGB, ABI2, RELA, RCN2, RARA, ST3GAL4, ACSS2, DEFB103B, ST2, ARL6IP1, TLR9, HNF1A, SOX18, PIEZO1, SYT1, STK11, SAMD9, STAT6, PMPCA, SNAI2, MPRIP, SRI, KDM6B, MAGI2, SPG7, PALLD, WDR1, SIGLEC1, SELENOS, TTR, TXNIP, FOLR2, LIF, CAPN1, CR1, CCDC80, RPSA, CAST, DAB2IP, CASP9, MIR142, MIR652, MIR134, CPB2, COX8A, MSTN, CD28, COMT, GHRHR, PRSS55, CD69, GJA5, NCF1, CXADRP1, UCA1, DAB2, LGALS3BP, MIR150, OSCP1, GAST, HLP, FGFR1, SLCO6A1, BCR, MIR375, MCAM, IRGM, MC1R, FGG, SMAD7, SMAD3, HOTAIR, CSF1, MAA, BMP7, BRCA1, C1QBP, C3, MIR181A2, LRP2, LPP, CACNA1C, MIR152, MIR10B, CDH5, GLO1, ITGAV, IGFBP3, IGFBP1, FFAR4, GUCY1A1, IRF8, CNN2, CMKLR1, HSPA1A, GZMB, CHRNA4, CISH, MIA3, NEXN-AS1, CLIC1, HDAC1, MIR146B, NANOS3, GSTK1, HNF4A, IRF2BP2, NRG1, TICAM2, HMGA1, CCL4L1, CHGA, IGFBP7, CNR1, COL15A1, GOT2, COL4A1, MIRLET7B, CDKN1A, IRS1, GPER1, IRF5, INPP5D, CXCL10, CDKN1B, C1QL3, CXCR2, IL6R, IL5, NR3C1, CXCL2, MIR499A, CEL, CTSC, CNP, GPIHBP1, NAXE, ACTRT1, CCR3, RNU6-392P, JAG1, NPR3, NPPA, DSPP, AGTR2, CCN3, MIR23B, LARP1BP2, GFM1, CYP7A1, TSLP, CYP1A2, CYP1A1, AKT2, MIR29B1, EDA, MIR135B, LPAR1, SESN2, NEXN, CTSL, MIR25, HAVCR2, CYP19A1, ACAN, CYP2J2, PAEP, PCSK6, HHIPL1, LOC107985770, MIR133B, DNAH8, ADK, KISS1R, SPZ1, OSM, ADRB2, ACCS, DNASE1, ATN1, DSC1, ARID5B, OGN, OGG1, RN7SL263P, ADTRP, DEFB103A, MIR148B, APC, H3P31, PDGFB, POSTN, DBP, KLRC4-KLRK1, C1QTNF6, TNFRSF13C, MSRA, ABCA4, SNHG16, FBN2, FCGR2B, ADA, CTF1, ADAM33, FGB, TMED7-TICAM2, MIR30E, MMP7, FOXO4, EARS2, FGF1, OSCAR, COL18A1, PLIN5, MYH11, BIRC3, EPHA1, EPO, ERG, F7, MIR99A, RHOA, ACP3, NEU1, ORMDL3, MIR206, ARR3, F13A1, MIR29B2, FABP2, LINC-ROR, MIR490, MIR17HG, SLC39A1, SIGLEC9, ARHGEF26, IL17B, PDCD4, MIR370, TES, MIR330, RNU1-1, MIR338, HAVCR1, HBP1, MIR378A, MIR335, MIR449A, H4C15, FETUB, PCOLCE2, MIR362, MIR489, MIR18B, MIR431, MIR328, HNP1, MIR451A, MIR410, PLA2G2D, FBXO3, MIR342, CLEC4E, FBXO8, DDAH1, APPL1, MIR876, ADAMTS5, CIT, WDHD1, GATD3B, HIF1A-AS1, SLC2A6, MIR4463, SNF8, COPE, P2RX5-TAX1BP3, MICOS10-NBL1, ACOT7, MGLL, MICA, CARD8, OCLN, MIR3188, SIRT2, AAA1, MIR664A, DEFB4B, CASC11, P2RX2, MYH15, HOXA-AS3, LOC102724197, CDR1-AS, MIR222HG, CCR9, CAMKK2, H3P28, CTCF, H3P23, LNCRNA-ATB, DCTN6, LINC02605, TRAF3IP2, NES, MTHFD2, CYSLTR1, MASCRNA, ESM1, FRS2, GSC-DT, LILRB1, USP20, SIRT1-AS, MALT1, BRD8, EBNA1BP2, LILRB4, ABHD2, KERA, C20orf181, STAB1, TMEM98, TLX1NB, MIR455, CARHSP1, PATZ1, MIR486-1, CLEC5A, DAPK2, BACE1, FIQTL1, MIR505, EID1, PANX1, ZNF318, GCA, LMOD1, BAMBI, MIR519D, PART1, MIR494, POT1, CLIC4, WWTR1, PNKD, ADGRA2, CHMP2B, MIR492, LEF1-AS1, ECSCR, NORAD, CES3, MLC1, PSS, NBEAL2, PACS2, MCF2L, SASH1, MIR758, SFTPA2, POTEF, KCNH4, TRAM1, SRRM2, DDAH2, CLCF1, NNT, MIR647, MIR612, MMD, MIR575, NME1-NME2, PIK3R5, SLC16A8, ATP6V0A2, SFTPA1, RBMS3, STEAP4, BHLHE22, MIR302A, GPR119, CD248, JPH3, STRIP2, MAGEE2, MAGEC3, KCNH8, TMEM18, PODN, TNFAIP8L1, SLC5A10, MRTFA, TDRD9, CIP2A, H4-16, PRAP1, MARK4, CCAR2, TADA1, SLAMF7, C1QTNF5, IL21, OSBPL8, OSBPL1A, CYGB, ZBTB46, TRPM6, SIRPA, SLC2A4RG, PAQR7, PDGFC, MARCHF10, TRPV5, LTB4R2, LINC00599, CYP26B1, AMOT, SLC2A9, SUCNR1, BTLA, BPIFB4, SERPINA12, SPHK2, LGALS14, ANKS1B, PNO1, PDIK1L, IL34, FAM20C, NRG4, PWAR1, AKR1B10, TBX20, SIGIRR, IL17F, EGLN3, JAM3, RETNLB, MAK16, METRN, KCTD15, LCOR, AKT1S1, MINDY4, FA2H, CCDC8, TAF3, SLC52A2, MFRP, UBE2Z, ARHGAP24, PLA2G12A, COLEC12, APOL6, HMBOX1, TNFAIP8L2, RHBDF2, SPX, PPP1R3B, PNPLA3, NAA25, AUNIP, CREB3L3, SENP2, ROBO3, MUC16, CLEC6A, CPAT1, ARHGAP18, CACNA2D4, GGTLC1, CIDEC, SAMSN1, SLC39A8, WDR20, CARD9, RSAD2, PINK1, DPP9, SAMD1, WNT3A, ARHGEF28, DCLK3, FBRS, ABCC11, HSH2D, P2RY12, ORAI1, AGXT2, ADAMTS18, PAOX, MLKL, INSIG2, MIR182, TRAT1, MIR18A, ADM2, MIR15A, MIR149, MIR148A, MLXIPL, MIR140, MIR136, MIR127, EGFL7, MIR185, MIR125A, PLEKHO1, GP6, MZB1, TLR8, MIR107, MIR100, SIRT7, LINC01194, HACD4, SNX19, MIR183, IL22, UFM1, HILPDA, IGLV2-18, IGKV2D-29, MIR29C, KLF15, PYCARD, BRD7, MIR296, CD274, MIR28, ICOS, MIR27B, MIR23A, MIR188, OSGIN1, MIR224, MIR22, MIR214, MIR205, PSAT1, MIR199A2, SLC40A1, MIR199A1, MIR192, PLA2G3, C1QTNF12, TBPL2, APOBR, SOX6, MOCOS, ATG16L1, HCCAT5, CLEC9A, RCBTB1, CSGALNACT2, H19, SGMS1, UTS2B, FNDC5, SMPD3, NSUN5, TESC, LRP2BP, LINC00305, ADCY10, ZEB1-AS1, CISD1, MPEG1, HYLS1, UNC5B, KMT2E, ZC4H2, TET3, SCARA5, S100A7A, ENHO, KRT20, NBAS, INPP5K, TM6SF2, PTOV1, P2RY13, FCRL6, NANS, GOLGA6A, ACER2, ARSI, TOLLIP, SMOX, CDKAL1, LINC00299, ARMH1, CCHCR1, BRINP3, DLL4, NANOS2, ENPP7, PGPEP1, MMS, DPP8, CLEC4D, C1QTNF1, A2M, HEXIM1, IGHG3, ITPR2, ITPR1, ITIH4, ITGB5, ITGB4, ITGAM, ITGAL, ITGAE, ITGAD, ITGA5, ITGA2, IRF3, IRF2, ILK, IL11, IL7, IL2, ITPR3, IVD, JUN, LAD1, LIPE, LECT2, LDHA, LBR, LBP, LAIR1, LAG3, KPNA4, JUNB, KPNA3, KLK1, KISS1, KCNMB1, KCNJ11, JUP, JUND, IGSF1, IGHE, LMNB1, IGFBP5, HAL, GYPC, GTS, GTF3A, GSTM2, GSR, GPX4, FFAR1, GPR39, LPAR4, CXCR3, GP1BA, GOLGB1, GLRX, GLI2, GCLM, GFER, HAS2, HAS3, HCRT, HRH2, IGF2R, IFI27, IDH2, ID1, HSPA2, HSF1, HES1, HPX, HDC, HOXB9, HOXA1, HNRNPU, FOXA3, HLA-B, HLA-A, HDLBP, FADS1, LRP5, NR6A1, NDUFA2, ROR2, YBX1, NRF1, SLC11A2, PNP, NOTCH2, NOP2, NNMT, NME2, NINJ2, NGFR, NFKBIA, NFKB2, NFE2, NFIA, NF1, NEDD4, DDR2, OPRK1, SLC22A18, CNTN3, PCSK1, PCP4, PCOS1, PCOLCE, PCNA, PCDH8, PC, PAK1, OSBP, PAH, PEBP1, P2RY1, P2RX5, P2RX4, P2RX3, P2RX1, NDUFAB1, NCF2, LSAMP, NBN, CXCL9, CD99, MEF2C, MEF2A, MECP2, MDM2, MDK, SMCP, CD46, MCM3, MCL1, MC4R, SMAD1, LYZ, CD180, LUM, LTB, MAP3K10, KMT2A, MMP10, RNR1, NBL1, NAGLU, MYH9, MUC1, TRNL2, TRNL1, MTRR, CYTB, MMP11, MTAP, MT1F, MST1R, MST1, ABCC1, MNDA, MMP17, GDF2, GCKR, PDE1A, BDKRB2, CD86, CD80, CD27, MS4A1, CD19, KRIT1, RUNX2, CAPG, CANX, CALU, CALR, CA1, SERPING1, BLM, PRDM1, BIK, CEACAM1, CD70, CD74, CD81, LYST, CORT, COL1A2, CCR8, CCR6, CMA1, CLCN2, CHRNA5, CEBPB, CDC42, CEBPA, CDKN1C, CDK9, CDK5, CDK2, CDH13, CDH1, BDNF, CCND1, CPOX, BAX, ALOX12, ABCD1, ALCAM, AHCY, NR0B1, AGRP, AP2A1, ADRB3, ADH5, ADCYAP1, ADCY9, ADCY8, ACTA2, ACP5, ACP1, ABR, SERPINA3, ALPL, AMH, ANXA2, ATF4, AZU1, AZGP1, ATP7A, ATP2B1, ATOX1, ATOH1, ATIC, ASS1, APEX1, ARSL, STS, ARSB, KLK3, APOC2, APOC1, XIAP, CLDN7, CPT2, GCK, TYMP, FDPS, FBLN2, FAT1, FAP, FABP3, F12, F11, ETS1, ERN1, EREG, EPAS1, EMP1, MARK2, ELAVL1, EFNB1, EEF1B2P2, EDNRB, FGA, FGF12, FGFR4, FUT7, GCGR, GATA4, GATA2, LRRC32, FYN, ACKR1, FUT8, FUT3, FHL2, FTH1, NR5A2, FOSB, FMOD, FLT4, FOXM1, VEGFD, EDN3, EBF1, CREB1, DUSP5, CYP2D6, CYP2C8, CYP2C19, CYLD, CTSG, CTSB, CTPS1, CTNND1, CTBS, NKX2-5, CSF3, CSF2RB, CSF1R, CRYGD, CRY1, CRMP1, ATF2, CYP2E1, CYP3A4, CYP3A5, DHCR24, RCAN1, DNMT3B, DMRT1, DMD, DLD, DIO2, NQO1, DHCR7, CYP26A1, DEFB4A, DDIT3, GADD45A, DBN1, BRINP1, DAG1, CYP27B1, PCSK5, PDE4A, PDLIM5, DENR, SPHK1, VNN1, ALKBH1, HCAR3, CCN5, NRP1, SUCLA2, TNFRSF11A, TNFRSF6B, TNFRSF14, ADAM9, TNFSF13, TNFRSF25, DGAT1, VAMP8, DYNLL1, TP63, SGPL1, TIMELESS, PHOX2B, ARHGEF1, MAGI1, ARHGEF2, OSMR, IL1RL1, TMSB10, HGS, PDCD5, P2RX6, TRPA1, SLC16A3, CLDN1, PSTPIP2, ARTN, CH25H, KALRN, SELENBP1, PLA2G4C, CYP4F2, COPB2, HAT1, HMGA2, AAAS, KMT2D, ADAM12, ARHGEF5, SCG2, FZD5, LEPQTL1, NPHS2, ZNF202, ZNF148, BEST1, VIP, VAV2, VAV1, VARS1, UTRN, SLC7A5, NRIP1, GATD3A, H4C8, PIK3R3, PPM1D, NR0B2, H4C14, H4C13, H4C5, H4C2, H4C3, KDM5D, H4C11, H4C12, H4C6, H4C4, H4C1, BRAP, H4C9, PDLIM7, GPR55, SUMO1, SH2B3, TLR6, LANCL1, TCIRG1, STUB1, FSTL3, SPRY1, RABEPK, KLRG1, MPZL2, PSME3, PRMT3, SORBS3, ATP6AP2, EBI3, ZNF263, IL18BP, DNM1L, MICU1, DLC1, PEMT, KAT5, TXNRD2, CXCL13, HTATIP2, PRDX4, PROCR, ZNRD2, ATG7, SEMA4D, SPON2, STK25, CREB3, CAP1, FST, VAV3, C1D, LYPLA1, PARP3, HDAC6, SLIT2, AKT3, TRAF4, AKAP12, NR1D1, GOSR2, H6PD, CXCL14, PTGES, NPEPPS, ADAMTS1, ATG5, CHST3, PCYT1B, GSTO1, ABCG2, FADS2, COX5A, PPIG, PHF14, KDM4A, ECE2, MAFB, SCO2, KCNE2, RCE1, FGF19, MVP, HS3ST1, DLEC1, MFN2, SEMA3E, ELMO1, TRIM14, MAML1, MTSS1, ATG13, KMT2B, DOCK4, UMOD, TYROBP, PDE9A, PTHLH, RFX5, RFX1, RFC2, RFC1, REG1A, RASGRF1, RASA1, RARB, RAG2, RAC2, RAC1, NECTIN1, PVR, PTPRF, PTPN11, PTPN6, PTPN2, RGS1, RGS2, RGS3, SAT1, CCL20, CCL17, CCL8, CCL7, CCL1, SCP2, SCN7A, TSPAN31, RGS7, SAA3P, S100A7, RPS6KA1, RNASE3, RNASE1, RLN2, RHEBP1, PTN, PTGIS, CCL25, PTGER4, POU2F1, POMC, PODXL, PRRX1, SERPINF2, PLAG1, PITX3, PITX2, PIP, SERPINB9, SERPINA1, PGC, SLC26A4, PDK4, PDK1, PDGFRB, PDGFA, MED1, PPP1R1A, PPP2R2A, RELN, PTGER3, PTBP1, PSMD9, PSMD7, PSMC5, PSMB8, PSMA6, PROX1, PTPA, PRNP, PRL, MAP2K7, MAPK9, PRKCB, PRKCA, PREP, CCL23, SDC1, TXNRD1, AURKA, THM, TGFBI, TGFB2, TGFB1I1, TFRC, TFAM, TEK, TDO2, TRBV20OR9-2, TCF15, ZEB1, TBXAS1, TBXA2R, TAZ, MAP3K7, TAC1, ABCC8, THY1, TLR1, TNFAIP6, TRAF1, TWIST1, TSC1, TRPC6, TRPC5, TRPC1, TRL-TAG1-1, TRAF2, HSP90B1, TNNI3, TPT1, TPO, TPM2, TPI1, TPH1, TP73, TNNT2, SULT2A1, STIM1, SDC4, NAT2, SMARCA4, SLPI, SLC22A1, SLC12A3, SLC8A1, SLC6A8, SLC6A4, SLC3A2, SKP2, ST8SIA1, ST6GAL1, SHC1, SGCB, SRSF2, SRSF1, SFRP4, SELL, SMN1, SMN2, SMPD2, SRY, STAR, ST14, SSTR2, SST, SSRP1, SSBP1, TRIM21, SRC, SMS, SPI1, SPARC, SP1, SOS1, SOD3, SNTB2, SNAI1, H3P10
-
Common Green Bottle Fly
Wikipedia
A new antimicrobial agent was isolated from L. sericata secretions and patented under the name Seraticin . [27] Efforts are geared toward making medical professionals more familiar to the current techniques. [28] Like many other ectoparasites , L. sericata has a huge economic impact on farmers, so many studies and research projects have been put in place since the late 1980s to help farmers reduce their impact.
- Bone Fracture Wikipedia
-
Internet Addiction Disorder
Wikipedia
They resort to virtual relationships and support to alleviate their loneliness. [27] [28] As a matter of fact, the most prevalent applications among Internet addicts are chat rooms, interactive games, instant messaging, or social media. [26] Some empirical studies reveal that conflict between parents and children and not living with mother significantly associated with IA after one year. [29] Protective factors such as quality communication between parents and children [30] and positive youth development [31] are demonstrated, in turn, to reduce the risk of IA. ... T=JS&PAGE=reference&D=psyc16&NEWS=N&AN=2020-30497-001. Accessed September 28, 2020. ^ Venkatesh, Viswanath; Sykes, Tracy Ann; Chan, Frank K. ... ISSN 0362-4331 . Retrieved 2018-02-28 . ^ Ko, Chih-Hung; Yen, Ju-Yu; Chen, Cheng-Sheng; Chen, Cheng-Chung; Yen, Cheng-Fang (February 2008). ... "Internet addiction among adolescents in Lebanon". Computers in Human Behavior . 28 (3): 1044–1053. doi : 10.1016/j.chb.2012.01.007 . ^ Bai, Ya-Mei; Lin, Chao-Cheng; Chen, Jen-Yeu (2001-10-01).
-
Polyploidy
Wikipedia
However multivalent pairing is common in many recently formed allopolyploids, so it is likely that the majority of meiotic stabilization occurs gradually through selection. [19] [21] Because pairing between homoeologous chromosomes is rare in established allopolyploids, they may benefit from fixed heterozygosity of homoeologous alleles. [28] In certain cases, such heterozygosity can have beneficial heterotic effects, either in terms of fitness in natural contexts or desirable traits in agricultural contexts. ... The Chromosomes (6th ed.). London: Chapman & Hall. p. 28. ^ Stebbins, G. L. (1950). "Chapter XII: The Karyotype". ... "Mechanisms of genomic rearrangements and gene expression changes in plant polyploids" . BioEssays . 28 (3): 240–252. doi : 10.1002/bies.20374 .
-
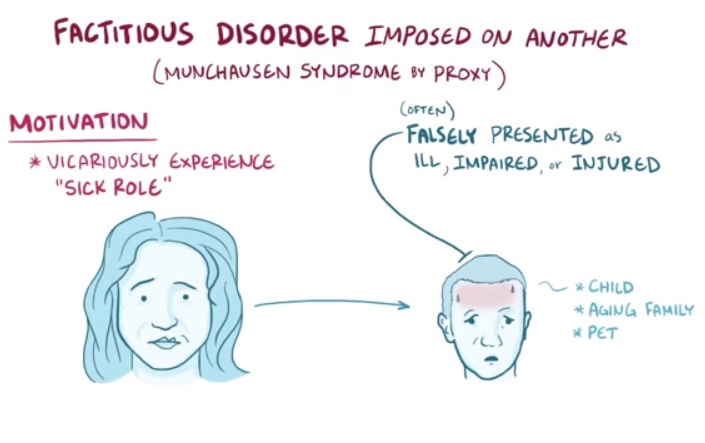
Factitious Disorder Imposed On Another
Wikipedia
., 2002) American Professional Society on the Abuse of Children proposed this term to diagnose the victim (child); the perpetrator (caregiver) would be diagnosed "factitious disorder by proxy"; MSbP would be retained as the name applied to the 'disorder' that contains these two elements, a diagnosis in the child and a diagnosis in the caretaker. [24] Induced Illness (Munchausen Syndrome by Proxy) (Ireland, 1999–2002) Department of Health and Children [17] Meadow's Syndrome (1984–1987) named after Roy Meadow. [25] This label, however, had already been in use since 1957 to describe a completely unrelated and rare form of cardiomyopathy . [26] Polle Syndrome (1977–1984) coined by Burman and Stevens, from the then-common belief that Baron Münchhausen's second wife gave birth to a daughter named Polle during their marriage. [27] [28] The baron declared that the baby was not his, and the child died from "seizures" at the age of 10 months. ... Merck Manuals Professional Edition . Retrieved 28 April 2019 . ^ a b c d e f g Criddle, L. (2010).
-
Asthma
Wikipedia
Asthma is characterized by recurrent episodes of wheezing , shortness of breath , chest tightness , and coughing . [23] Sputum may be produced from the lung by coughing but is often hard to bring up. [24] During recovery from an asthma attack (exacerbation), it may appear pus-like due to high levels of white blood cells called eosinophils . [25] Symptoms are usually worse at night and in the early morning or in response to exercise or cold air. [26] Some people with asthma rarely experience symptoms, usually in response to triggers, whereas others may react frequently and readily and experience persistent symptoms. [27] Associated conditions A number of other health conditions occur more frequently in people with asthma, including gastro-esophageal reflux disease (GERD), rhinosinusitis , and obstructive sleep apnea . [28] Psychological disorders are also more common, [29] with anxiety disorders occurring in between 16–52% and mood disorders in 14–41%. [30] It is not known whether asthma causes psychological problems or psychological problems lead to asthma. [31] Those with asthma, especially if it is poorly controlled, are at increased risk for radiocontrast reactions. [32] Cavities occur more often in people with asthma. [33] This may be related to the effect of beta 2 agonists decreasing saliva. [34] These medications may also increase the risk of dental erosions . [34] Causes Asthma is caused by a combination of complex and incompletely understood environmental and genetic interactions. [4] [35] These influence both its severity and its responsiveness to treatment. [36] It is believed that the recent increased rates of asthma are due to changing epigenetics ( heritable factors other than those related to the DNA sequence ) and a changing living environment. [37] Asthma that starts before the age of 12 years old is more likely due to genetic influence, while onset after age 12 is more likely due to environmental influence. [38] Environmental See also: Asthma-related microbes Many environmental factors have been associated with asthma's development and exacerbation, including, allergens, air pollution, and other environmental chemicals. [39] Smoking during pregnancy and after delivery is associated with a greater risk of asthma-like symptoms. [40] Low air quality from environmental factors such as traffic pollution or high ozone levels [41] has been associated with both asthma development and increased asthma severity. [42] Over half of cases in children in the United States occur in areas when air quality is below the EPA standards. [43] Low air quality is more common in low-income and minority communities. [44] Exposure to indoor volatile organic compounds may be a trigger for asthma; formaldehyde exposure, for example, has a positive association. [45] Phthalates in certain types of PVC are associated with asthma in both children and adults. [46] [47] While exposure to pesticides is linked to the development of asthma, a cause and effect relationship has yet to be established. [48] [49] The majority of the evidence does not support a causal role between acetaminophen (paracetamol) or antibiotic use and asthma. [50] [51] A 2014 systematic review found that the association between acetaminophen use and asthma disappeared when respiratory infections were taken into account. [52] Acetaminophen use by a mother during pregnancy is also associated with an increased risk of the child developing asthma. [53] Maternal psychological stress during pregnancy is a risk factor for the child to develop asthma. [54] Asthma is associated with exposure to indoor allergens. [55] Common indoor allergens include dust mites , cockroaches , animal dander (fragments of fur or feathers), and mold. [56] [57] Efforts to decrease dust mites have been found to be ineffective on symptoms in sensitized subjects. [58] [59] Weak evidence suggests that efforts to decrease mold by repairing buildings may help improve asthma symptoms in adults. [60] Certain viral respiratory infections, such as respiratory syncytial virus and rhinovirus , [22] may increase the risk of developing asthma when acquired as young children. [61] Certain other infections, however, may decrease the risk. [22] Hygiene hypothesis The hygiene hypothesis attempts to explain the increased rates of asthma worldwide as a direct and unintended result of reduced exposure, during childhood, to non-pathogenic bacteria and viruses. [62] [63] It has been proposed that the reduced exposure to bacteria and viruses is due, in part, to increased cleanliness and decreased family size in modern societies. [64] Exposure to bacterial endotoxin in early childhood may prevent the development of asthma, but exposure at an older age may provoke bronchoconstriction. [65] Evidence supporting the hygiene hypothesis includes lower rates of asthma on farms and in households with pets. [64] Use of antibiotics in early life has been linked to the development of asthma. [66] Also, delivery via caesarean section is associated with an increased risk (estimated at 20–80%) of asthma – this increased risk is attributed to the lack of healthy bacterial colonization that the newborn would have acquired from passage through the birth canal. [67] [68] There is a link between asthma and the degree of affluence which may be related to the hygiene hypothesis as less affluent individuals often have more exposure to bacteria and viruses. [69] Genetic CD14-endotoxin interaction based on CD14 SNP C-159T [70] Endotoxin levels CC genotype TT genotype High exposure Low risk High risk Low exposure High risk Low risk Family history is a risk factor for asthma, with many different genes being implicated. [71] If one identical twin is affected, the probability of the other having the disease is approximately 25%. [71] By the end of 2005, 25 genes had been associated with asthma in six or more separate populations, including GSTM1 , IL10 , CTLA-4 , SPINK5 , LTC4S , IL4R and ADAM33 , among others. [72] Many of these genes are related to the immune system or modulating inflammation.
-
Phobia
Wikipedia
As the phobic person approaches a feared stimulus, anxiety levels increase, and the degree to which the person perceives they might escape from the stimulus affects the intensity of fear in instances such as riding an elevator (e.g. anxiety increases at the midway point between floors and decreases when the floor is reached and the doors open). [28] Treatments [ edit ] There are various methods used to treat phobias.
-
Benign Paroxysmal Positional Vertigo
Wikipedia
Treatment is therefore geared toward moving the canalith from the lateral canal into the vestibule. [28] The roll maneuver or its variations are used, and involve rolling the person 360 degrees in a series of steps to reposition the particles. [1] [29] This maneuver is generally performed by a trained clinician who begins seated at the head of the examination table with the person supine [30] There are four stages, each a minute apart, and at the third position the horizontal canal is oriented in a vertical position with the person's neck flexed and on forearm and elbows. [30] When all four stages are completed, the head roll test is repeated, and if negative, treatment ceases. [30] Medications [ edit ] Medical treatment with anti-vertigo medications may be considered in acute, severe exacerbation of BPPV, but in most cases are not indicated.
-
Hyperglycemia
Wikipedia
Severe hyperglycemia can be treated with oral hypoglycemic therapy and lifestyle modification. [28] Replacing white bread with whole wheat may help reduce hyperglycemia In diabetes mellitus (by far the most common cause of chronic hyperglycemia), treatment aims at maintaining blood glucose at a level as close to normal as possible, in order to avoid serious long-term complications.INS, PRKCB, INSR, GCK, IL6, HNF1A, NOS3, LEP, GCG, NFE2L2, CCL2, AGER, ADIPOQ, PDX1, PTGS2, LEPR, IRS2, SP1, GPX1, NQO1, CD40, SIM1, HSD11B1, FBN1, FCGR3B, GLP1R, HMGA1, PRKAR1A, CD163, AKR1B1, PRDX4, ICAM1, COL3A1, IL1B, ITGAM, MMP9, PPARGC1A, AVP, PRKCZ, HCRT, GRK2, ADIPOR1, LDHA, PCSK1, HSPD1, HTR2A, PRNP, CBL, CYP11A1, EPAS1, STAT3, HRH3, PKLR, PPARG, CNTF, DRD1, PLA2G4A, ADA, ADRB1, HNF4A, HCRTR2, GIPR, ABCC8, KCNJ11, PIK3R1, PAX4, MC4R, NEUROD1, GATA6, APPL1, PLAGL1, ACAT1, KLF11, SLC2A4, IL10, PTF1A, IRS1, HYMAI, LMNA, ZFP57, CRK, SI, ST3GAL4, ITPR3, AHSA1, TXNIP, RFX6, SLC2A1, SLC5A2, MAPK14, ATN1, IGF1, DPP4, GIP, FGF21, GCGR, GABPA, PIK3CD, PTPN22, PIK3CB, POLDIP2, PRKAA1, FOXO1, PRKAA2, PIK3CA, PHKG2, GPT, EHMT1, ARMC5, MAPK1, RNF19A, HIF1A, REN, ROS1, PIK3CG, IAPP, CLCNKB, SIRT1, BLK, MGAM, TNF, ALB, AIMP2, VEGFA, ZMPSTE24, TCF7L2, TLR4, LRPPRC, TGFB1, AKT1, CEL, EIF2AK3, TP53, INS-IGF2, GRAP2, ZGLP1, SLC5A1, CAT, DECR1, PRKCA, SERPINE1, GH1, HMGB1, FN1, NLRP3, PRKAB1, PPARA, NOX4, ACE, DNM1L, MTOR, SOD2, AGT, WNK1, EPHB2, BDNF, SETD2, EPO, SYT1, CRP, APOE, SLC2A2, PTEN, GLO1, RELA, GCKR, HMOX1, IL18, VCAM1, THBS1, CYBB, POMC, GORASP1, GPBAR1, MOK, DAPK2, NFKB1, TXN, KDR, MIR146A, MIR126, UTRN, DENR, GSK3B, CRMP1, UCP2, RCBTB1, IL17A, CTNNB1, AGTR1, PDHX, TRPC6, GOLGA6A, VDR, MMP2, PON1, G6PC, FGF2, BCL2, FOXO3, APRT, PRRT2, ANGPT2, BGLAP, SLC17A5, SGK1, IFNG, CASP3, CD59, SIRT3, NFAT5, EDN1, MTNR1B, ALDH2, LOC102723407, EGR1, MAPK8, PTPN1, VWF, CXCL12, MFAP1, CD36, GYS1, ENPP1, PDK4, P2RX7, PARP1, SOCS3, MBL2, ACE2, PNPLA3, MAPT, LPL, ADCY5, TRIB3, BECN1, APOB, NR3C1, ROBO4, APOA1, AMY2A, IL22, GAL, TNFRSF11B, SERPINF1, FTO, MLXIPL, HK2, NUCB2, HSPA4, CXCL8, AKT2, NGF, HGF, IGF1R, FAM3B, MTHFR, IL1RN, IL4, FFAR1, SOD1, MIR223, MALAT1, CDH5, FGF1, FASN, CREM, PTK2B, ROCK1, SHBG, TDGF1P3, BMP7, F3, MAPK3, TGM2, ESR1, DIANPH, MIR34A, FST, ASPG, RAC1, SLC30A8, SELP, STK11, TLR2, ATF3, APP, VDAC1, PLA2G1B, ARG1, PLG, APOC3, TTR, PPARD, CASP1, SELE, HPSE, SELENOP, NSA2, KLF2, MST1, STAM, NR3C2, CCL5, SCT, PECAM1, SLC30A10, LOH19CR1, UNC13B, TNFSF10, SNAP25, LGALS3, PDE5A, SMN2, SMN1, SPP1, SREBF1, APOM, SMAD3, SREBF2, SLC5A3, MBD2, SST, MDM2, TCF21, NOS1, MYD88, KL, TKT, LONP1, PAX6, PC, BRD7, RAPGEF5, CRISP2, PCK2, TRPV1, TRPC3, PCNA, TSC2, MCAT, ABCG2, SERPINA1, HIPK2, REG3A, PRKCD, KLRK1, YWHAZ, NEU1, MMRN1, SLC39A7, RPS19, CXCR4, KDM1A, TEK, TFAM, PTH, NOS2, WFS1, RENBP, NOTCH1, NPY, NAMPT, SMUG1, ABCA1, DLK1, KMT5AP1, ESR2, CRY1, CSF1, DBP, HSPA12A, NRK, PRSS55, DNMT1, DRD2, RCAN1, CRTC2, PLB1, EGFR, SLC30A7, ENO2, FABP4, HHIP, FOXM1, FOS, FOSB, GFAP, GFPT1, SPZ1, GJA1, GLS, SETD7, GYS2, HBG2, CORO7, HMGCR, HOXD13, CREB1, COMP, MIR140, CHRM3, ACHE, ACTB, ADORA2B, ADRB3, AGTR2, KLRC4-KLRK1, FOXO6, C20orf181, ALOX15, ANPEP, AQP7, AQP9, ATIC, ATM, MIR375, BCHE, BNIP3, BTC, TSPO, VPS51, MIR26B, CAV1, CAV3, MIR24-1, CD86, MIR155, CDK4, CEBPA, FOXN3, P2RY12, DUSP1, HSF1, FAM3A, IDE, IGF2, G6PC2, IGFBP2, CXCL10, JUN, NOD2, HSPG2, JUNB, HSPA5, JUND, HSP90AA1, IL1R1, IL1A, MIR29A, SLC38A4, RACK1, SYCP2, MIR106B, MICU1, MIR23A, AGGF1, PNPO, IL21, MIR106A, TRPM7, ABCC4, MIR27A, RETSAT, CD226, LPCAT3, MIR29C, RHOT1, MIRLET7C, MIR301A, RASGRP1, CTDSP2, FBXW7, PPIF, NLRC4, VN1R17P, PTPRU, MIRLET7B, STAP2, MIR206, IFI30, MIR129-2, TRPV4, MIR132, CCT2, CASZ1, SLC35A1, SCPEP1, CYP2W1, PID1, ATG7, MIR144, DEAF1, MIR145, SEMA3C, MIR107, MIR150, FAM3C, MIR152, NCOA2, GER, VAT1, MIR17, MIR182, PDPN, THRAP3, MAD2L2, MIR200B, IGF2BP2, CXCR6, MIR200C, HCN4, XRCC6P5, GFPT2, MIR4454, SLC16A4, SLC16A3, PGR-AS1, P2RX6, LPAR2, SLC12A9, POLD4, KLF4, P2RX5-TAX1BP3, CHPT1, XPR1, MTA2, DIP, S1PR2, USP14, SNAP23, LOC102723971, F2RL3, TRPA1, DALIR, PERCC1, CST12P, SGPL1, SPHK1, RN7SL263P, ACKR3, RPTOR, ARHGEF7, KAT2B, H3P5, PROM1, TRAP, RETN, GPR166P, DCAF1, MIR503, MIR451A, GDF15, PTBP2, PRDX6, MIR423, HDAC9, POLE4, CTDSP1, ZDHHC7, BMS1, MIR135B, DDIT4, RNF10, DELYQ11, MIR486-1, GAL3ST1, ATG5, NCF1, PICK1, GGTLC5P, MAP4K4, ANGPTL8, MIR33B, MIR592, GGTLC3, POTEF, GGT2, SLC22A8, CCR2, GGTLC4P, SIX2, SMIM1, MAFA, GALP, NEXN, HSPB8, IL33, WNT3A, SIGLEC7, INTU, SERP1, QRFPR, ORAI1, HPGDS, BHLHE22, CNDP1, COX4I2, DESI1, DNER, FOXP2, FBXO8, FBXO2, EGLN3, MRAP2, MARCHF3, FN3K, KIFBP, NPVF, SNAP47, SNED1, MRGPRX3, GDE1, MRGPRX4, FTSJ3, MSI2, RPAIN, SIRT6, NES, DUOX2, TRPM5, CERS2, GGCT, A1CF, DUSP26, OBP2A, BRD9, HMCN1, HSPA14, ELOVL1, NEUROG3, TAS2R13, ASCC1, TVP23B, ABCG8, EFHD2, NOX5, GP6, ADIPOR2, NT5C3A, ASRGL1, NRN1, KLF15, HKDC1, SCUBE1, DACT1, KCNH6, TSC22D4, MCTS1, FLVCR1, SGSM3, RBM45, GPR151, NGDN, KRT20, METAP2, CAPN10, PSIP1, PTGDR2, GPRC6A, INPP5F, SIRT2, CHMP2B, DKK1, ERRFI1, P2RX2, NT5C2, UNC13A, SLC25A19, CADM2, PCSK9, EHMT2, LGR6, MTMR11, MRGPRX1, RAB10, C1QL1, OR10A4, ZFP69, QRFP, PWAR4, GPR142, ZACN, ENHO, FBRS, SRSF10, ELOVL5, MCF2L2, SLC24A3, SIK1, PTRH1, TRIM69, SH2B1, SRXN1, SIRPA, PART1, EMCN, TXN2, FTSJ1, FGFBP3, NRG4, TICAM1, LY96, LUC7L3, PROK2, ATG4B, LPAR3, DDAH2, PES1, SLC35G1, ANGPTL2, OXER1, GLIS3, SIRT5, NEDD4L, DUOX1, CENPX, ZBTB7C, PHLPP1, TBC1D1, SEMA6A, RPS6KB1, RIPK2, GCLC, GATA4, GCH1, GDF2, MSTN, GDNF, GFER, GGT1, GHR, GHSR, GLDC, GALNT2, GLUL, CXCR3, GPER1, GPR42, GPX5, GSTT1, GUSB, HAS2, HBB, GAS6, GALR1, TNFRSF14, F2, EEF1A2, EGF, ELAVL1, ENO1, EP300, EPHX2, EPOR, ESRRG, EZH2, FAAH, GAD1, FABP1, FANCC, FGB, FGL1, FGR, FLT1, FMO3, NR5A1, GAST, HCLS1, HCRTR1, HLA-B, KRAS, IL13, FOXK2, INSRR, IRF3, ITGA2, JAK2, KCNH2, KCNQ1, KNG1, LDLR, HLA-DMA, LECT2, LGALS1, LIPE, LOX, LRP5, LTA, LTBR, SMAD2, MAP2, IL7R, IL2RB, IL2, IKBKB, HLA-DMB, HLA-DRB1, NR4A1, SLC29A2, HNRNPA1, TLX2, HPRT1, AGFG1, PRMT1, HES1, HSPA1A, HSPA1B, HTR2C, IARS1, IRF8, IGFBP1, IGFBP3, IGFBP7, CCN1, EDNRA, S1PR3, S1PR1, CCND1, AQP5, AR, ARF6, ARRB1, ARRB2, ARSA, ASAH1, ATP2A3, BAX, HCN2, CD33, BMP4, BRS3, KLF9, C5AR1, CACNA1D, CAPN1, CASP8, CASR, RUNX2, AQP3, KLK3, APOA4, XIAP, ACACA, ACVRL1, ADCYAP1, PLIN2, ADK, ADM, ADRA1A, ADRA2B, JAG1, AHR, AHSG, AIF1, ALAD, ALDOB, ALK, ALX3, AMPD1, ANGPT1, ANXA5, MS4A1, ENTPD1, ECE1, DEFB1, CTLA4, CUX1, CYBA, CYC1, CYP1B1, CYP2E1, DAB2, DCN, DDIT3, TIMM8A, CD40LG, DIO2, DLD, DLG1, DMD, DMRT1, DYNC1H1, DUSP6, DUSP9, E2F1, CCN2, VCAN, CSF2, CSF1R, CD44, CD68, LRBA, CDC25C, CDC42, CDK2, CDK5, CDKN1A, CEACAM5, CMA1, CMKLR1, CNR1, COPA, COX4I1, CPT1A, CPT2, CREBBP, CRH, CRYZ, MDM4, MEFV, MAP3K5, SULT1E1, SNCA, SOAT1, SORD, SPARC, SRC, SSTR4, ST13, STAR, STAT5A, SUV39H1, TFPI, SYCP1, TAC1, TAT, TBX1, TBXA2R, TCF3, ZEB1, PPP1R11, TF, SMO, SNAI2, SLPI, SLC20A1, S100B, ACSM3, SAT1, SCD, SCN10A, SCP2, CCL3, CCL18, CXCL5, CX3CL1, SDC2, SDC4, SIX3, SLC1A3, SLC2A3, SLC6A6, SLC9A1, SLC16A1, SLC19A1, TFE3, TFRC, SORT1, ULK1, VIP, BEST1, CLIP2, YY1, CNBP, AIR, MLRL, FZD4, PLA2G6, STK24, TG, DOC2B, DGKD, APOL1, DEGS1, KHSRP, AOC3, NCOA1, RIPK1, TNFSF14, VIM, VEGFC, VEGFB, VDAC2, TGFB3, THBS2, KLF10, TIMP1, TIMP2, TIMP3, TLR1, TM7SF2, TMPRSS2, TNFRSF1A, TNFRSF1B, TPT1, TRH, UBE2V1, UCHL1, UCP3, UGCG, NR1H2, VASP, S100A8, S100A1, RAB8A, P2RX4, NTS, NR4A2, OGG1, OPA1, OPRK1, OPRM1, OSM, P2RX1, P2RX3, P2RX5, ABCB1, P2RY1, P2RY2, PAK1, PAWR, PAX3, PCK1, PDCD1, PDK1, PDR, DDR2, NRTN, SLC11A2, CCN3, MEN1, MFGE8, MIP, MITF, KMT2A, MMP3, MMP7, MMP16, MPG, MPO, ABCC1, MT2A, RNR2, MYC, MYLK, NCK1, NFE2, NFIL3, NFKBIA, PFKM, SLC25A3, CLIP1, RBP3, PTGDS, PTGS1, PTPN2, PTPN4, PTPN9, PTPRN, PTX3, RARA, RASGRF1, RBP4, PIM1, REG1A, RFC1, RHEB, BRD2, RORC, RPL10, RPL29, RPL36A, ABCA4, PTCH1, PROX1, MAP2K7, MAP2K6, PIN1, PIN4, PKM, PLA2G2A, PLAT, PLAUR, PLTP, PLXNA1, PML, PON2, POU1F1, CTSA, PPIA, PPID, PPP2CA, PTPA, PRKCI, PRKD1, MAPK7, H3P40