-
Primary Biliary Cholangitis
Wikipedia
Anti-gp210 antibodies are found in 47 percent of PBC patients. [26] [27] Anti-centromere antibodies often correlate with developing portal hypertension. [28] Anti-np62 [29] and anti-sp100 are also found in association with PBC. ... "Primary Biliary Cirrhosis: Getting a Diagnosis" (online) . At Home Magazine . Retrieved 28 July 2015 . ^ Jacoby A, Rannard A, Buck D, et al.IL12A, SPIB, IL12RB1, ABCC2, NOS2, MMEL1, TNFSF15, STAT4, POU2AF1, NFKB1, CLEC16A, IRF5, TNPO3, TYK2, IL7R, DENND1B, TNFRSF1A, SLC4A2, ABCB4, ALB, CD80, CXCL10, NFE2L2, TGFB1, LOXL2, IL12B, CXCL9, LOX, CXCR5, HIF1A, NOS3, KEAP1, SLC51A, ICAM3, MAPK14, CYGB, CDH5, SLC51B, IGFBP1, RELA, CCL5, UBD, ENTPD2, ABCG2, CD3D, NSA2, KRT7, IL4, PPARGC1A, HAMP, ACE, VEGFA, JAK2, SLCO1C1, MMP13, HMOX1, ACE2, DAG1, HHIP, CPEB1, MAS1, MAP3K14, MMP2, MMP3, PDE5A, MMP9, RPS6KB1, AGT, COL1A1, REN, CCN2, AQP4, HLA-DPB1, IL12RB2, IKZF3, HLA-DRA, IL16, GSDMB, IL21R, PRKCB, SYNGR1, ORMDL3, ZPBP2, TNFSF8, PAX3, DGKQ, MAPK8IP1P1, PLCL2, KANSL1-AS1, MAPK8IP1P2, PLCB1, RPL3, TP63, IL10, NSF, RAD51B, KANSL1, CD58, PSMG4, CYP21A2, ATXN2, LINC01100, LINC02820, LINC02210-CRHR1, SKIV2L, RPS6KA4, SP100, MAPT, MANBA, TSBP1-AS1, IL12A-AS1, ELMO1, LTBR, EIF4EP2, SH2B3, DLEU1, STAT1, MAPT-IT1, NAB1, TSBP1, DELEC1, MAPT-AS1, CCDC88B, ARHGAP31, PRICKLE1, GLI3, PSMD3, TMEM39A, LINC02210, SPPL2C, WNT3, SDK1, COL17A1, SLC22A23, SLC17A8, CNTN5, PRDM16, CTLA4, CDK12, GRIK1, HLA-DRB1, MED1, EIF4E, RBM45, ARID3A, TIMMDC1, STH, FBXL20, EXOC3L4, DLAT, RMI2, DDX6, TNF, VDR, IFNG, NUP210, PML, TLR4, FOXP3, CDKN1A, FGF19, IL17A, GPT, OGDH, IL1B, CD40LG, CXCL8, SQSTM1, CYP7A1, ABCB11, CDKN2A, HLA-DOA, PPARA, HK1, BCL2, DLST, PDC, ABCB1, TNFSF11, PDHX, TRIM21, NR1I2, ICAM1, SLPI, ATHS, MIR506, GGT1, CD14, SLC12A9, NAT10, HGF, GGTLC5P, HLA-DQA1, GGTLC3, GGT2, CD40, GGTLC4P, PDLIM3, CD19, PTPN22, IL2, ATRNL1, IL5, ASRGL1, ATN1, TBC1D9, MIR21, CCL27, DBT, ALPP, TGFBR2, XPR1, SULT2A1, SYCE1L, C4B_2, LOC102723407, LOC102724197, LOC102724971, TLR2, IL21, GPBAR1, SLC22A1, SLCO1B1, KHDRBS1, BTG3, NUP62, ABCC3, SOCS1, B3GAT1, DCTN4, GGTLC1, CDR3, NELFCD, KRT20, KLHL1, NR1H4, KLHL12, TP53, SLC10A2, H3P10, ESR2, LGALS3BP, HLA-A, CDKN1B, C4A, CYP3A4, C4B, CX3CR1, GTF2H1, POU2F1, MS4A1, CTSZ, GABPA, NRAS, CCR5, CD86, CXCR3, IL7, CD79A, FBL, ESR1, SLC10A1, HLA-DRB3, APOE, HNF4A, IL2RA, HSPD1, PPP1R2C, GLB1, GATM, PDCD1LG2, LRRC32, PNPLA3, MARCKSL1, FOXO3, FOXO1, WLS, FH, AGBL2, VTCN1, TNFAIP8L2, SLC25A1, POGLUT1, NR3C1, SIAE, IGF1, IFI27, SOST, TMED7, HMGB1, IL23A, HLA-DQB1, HLA-DPA1, HLA-B, TLR9, PDP1, COG6, RBFOX1, DPP8, GZMB, ITLN1, PRPF40A, WDR11, GSTM2, PRAM1, GSTM1, JPH3, JAM3, NQO1, NBPF3, MIR223, MIR505, CD1D, CD1C, DDR1, SPATA31A3, BRCA1, BHMT, BCR, BCL3, ATM, AREG, ABCC6, DEFB4B, FASLG, TMED7-TICAM2, FAS, APEX1, AIRE, ANXA11, ANXA2, ACTB, LINC02605, H3P23, MIR425, MIR210, DNMT1, CD28, DPP9, DNASE1, DMBT1, SP140L, TBX21, DBA2, DEFB4A, FCRL3, CTHRC1, DEFB1, LRG1, CYP2D6, COL9A3, CNGB1, CCR7, CHUK, CHRM3, CFTR, CD74, LINC01193, TICAM2, MIR155, MIR197, CD207, CD274, SCHIP1, POLR2G, SERPINA1, WNT2B, PRRC2A, PDK4, FOSL1, KMT2D, CDK2AP1, PDGFB, NR0B2, PDCD1, OPRM1, RNASET2, TNFRSF11B, DDR2, NOTCH1, TNFSF10, TNFSF9, TNFRSF11A, IL18R1, NFYA, MYD88, WASF1, ABCC1, HAP1, MIF, PIK3CA, VCAM1, EBAG9, UGT2B4, SOAT1, CXCL12, CCL24, SPINT1, SPP1, SRY, CCL20, CCL11, STAT3, SCTR, STAT5A, STAT5B, SERPINB3, TFF3, S100A12, RDX, TGM2, PTGS2, TLR3, PSMD9, PSMA4, PRF1, PPARG, TPMT, TNFSF4, CD99, MBL2, ICOS, TNFSF13B, ITGB2, ITGAV, RIPK3, ITGAL, PDZD2, ITGA5, ITGA1, ISG20, IL18, IL13, ICOSLG, SLC44A1, CLDN14, IL6, SS18L1, IL1RN, IL1A, INTU, AGO2, IL37, IGHG3, HPGDS, SLCO1B3, TRBV10-1, ABO, ITIH4, DCTN6, KLF4, KRT18, SLIT2, NTN1, SMAD3, HDAC9, KMT2B, SMAD2, CD180, LTA, LRP1, NR1I3, LPA, DGCR2, LORICRIN, EBI3, KLRG1, CALCOCO2, ABCC4, CCL26, LGALS3, CIB1, ZNRD2, LEP, GNLY, LAMP1, L1CAM, ABCA1
-
Dutch Elm Disease
Wikipedia
A policy of sanitary felling has kept losses in the city to an average of 1000 a year. [28] Elm was the most common tree in Paris from the 17th century; before the 1970s there were some 30,000 ormes parisiens . ... Contact (in French). Laval University. 28 (1). ^ CFIA annual pest survey report. 1999 Summary of Plant Quarantine Pest and Disease Situations in Canada (report available upon demand at the Canadian Food Inspection Agency: http://publications.gc.ca/site/eng/9.831610/publication.html ) ^ "Dutch Elm Disease" . ... Archived from the original (PDF) on 28 June 2007. ^ Screening European Elms for resistance to 'Ophiostoma novo-ulmi' ( Forest Science 2005) [2] ^ ‘Spanish Clones’ (Oct. 2013) resistantelms.co.uk ^ Δoκιμή ανθεκτικότητας ελληνικών γενoτύπων πεδινής φτελιάς (Ulmus minor) κατά της Oλλανδικής ασθένειας , Σ. ... Retrieved 16 November 2013 . ^ Coleman 2009 ^ 'First Genetically Modified Dutch Elm Trees Grown', unisci.com ^ resistantelms.co.uk, FAQ 'Disease Control' ^ Coleman 2009 , p. 17 ^ "The mid-Holocene Ulmus decline: a new way to evaluate the pathogen hypothesis" . Archived from the original on 28 September 2011 . Retrieved 4 November 2011 . ^ Oliver Rackham, The History of the Countryside (London 1986), pp. 242–243, 232 ^ Meulemans, M.; Parmentier, C. (1983).
-
Medial Knee Injuries
Wikipedia
This soft-tissue attachment can be reproduced with a suture anchor [28] placed 12.2 mm distal to the medial joint line (average location), directly medial to the anterior arm of the semimembranosus tibial attachment. [27] Once this aspect of the sMCL is secured to the suture anchor, the knee is put through range of motion testing by the physician to determine the "safe zone" of knee motion which is used during the first post-operative day rehabilitation (below). [27] Rehabilitation [ edit ] Nonoperative Rehabilitation As mentioned in the Nonoperative Treatment section, the principles of rehabilitation are to control swelling , protect the knee (bracing), reactivate the quadriceps muscle, and restore range of motion . ... The Physician and Sportsmedicine . 41 (3): 19–28. doi : 10.3810/psm.2013.09.2023 . ... PMID 20563561 . ^ Crawford, Duncan (28 January 2010). "Northern Ireland kneecapping victim 'shot four times ' " .
-
Chytridiomycosis
Wikipedia
Excessive shedding of skin is seen in most frog species affected by B. dendrobatidis . [6] These pieces of shed skin are described as opaque, gray-white, and tan. [6] Some of these patches of skin are also found adhered to the skin of the amphibians. [6] These signs of infection are often seen 12–15 days following exposure. [20] The most typical symptom of chytridiomycosis is thickening of skin, which promptly leads to the death of the infected individuals because those individuals cannot take in the proper nutrients, release toxins, or, in some cases, breathe. [6] Other common signs are reddening of the skin, convulsions, and a loss of righting reflex . [20] In tadpoles, B. dendrobatidis affects the mouthparts, where keratin is present, leading to abnormal feeding behaviors or discoloration of the mouth. [6] Research and impact [ edit ] The amphibian chytrid fungus appears to grow best between 17 and 25 °C, [21] and exposure of infected frogs to high temperatures can cure the frogs. [26] In nature, the more time individual frogs were found at temperatures above 25 °C, the less likely they were to be infected by the amphibian chytrid. [27] This may explain why chytridiomycosis-induced amphibian declines have occurred primarily at higher elevations and during cooler months. [28] Naturally produced cutaneous peptides can inhibit the growth of B. dendrobatidis when the infected amphibians are around temperatures near 10 °C (50 °F), allowing species like the northern leopard frog ( Rana pipiens ) to clear the infection in about 15% of cases. [29] Although many declines have been credited to the fungus B. dendrobatidis - although likely prematurely so in many cases [4] - some species resist the infection and some populations can survive with a low level of persistence of the disease. [30] In addition, some species that seem to resist the infection may actually harbor a nonpathogenic form of B. dendrobatidis . ... S2CID 4421285 . ^ "Amphibian 'apocalypse' caused by most destructive pathogen ever" . Animals . 2019-03-28 . Retrieved 2019-04-06 . ^ Briggs, Helen (29 March 2019). ... Retrieved 29 March 2019 . ^ Scheele, Ben C.; Pasmans, Frank; Skerratt, Lee F.; et al. (28 March 2019). "Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity" (PDF) .
- Alcohol And Pregnancy Wikipedia
-
Rheumatic Fever
Wikipedia
Duckett Jones, MD. [25] They have been periodically revised by the American Heart Association in collaboration with other groups. [26] According to revised Jones criteria, the diagnosis of rheumatic fever can be made when two of the major criteria, or one major criterion plus two minor criteria, are present along with evidence of streptococcal infection: elevated or rising antistreptolysin O titre [27] or Anti-DNase B . [28] [8] A recurrent episode is also diagnosed when three minor criteria are present. [29] Exceptions are chorea and indolent carditis , each of which by itself can indicate rheumatic fever. [30] [31] [32] An April 2013 review article in the Indian Journal of Medical Research stated that echocardiographic and Doppler (E & D) studies, despite some reservations about their utility, have identified a massive burden of rheumatic heart disease, which suggests the inadequacy of the 1992 Jones' criteria.MYOM2, CDKN2A, TP53, TNF, HLA-DRB1, MDM2, BMI1, PAGR1, RBM45, GALNS, GAST, COMMD3-BMI1, NPM1, MBL2, TLR2, IL6, CDK4, PTEN, TGFB1, CTLA4, IL10, ATM, IL17A, HLA-A, RUNX1T1, TBX3, FCN2, MASP2, EGFR, CBX7, HNF1A, TBX2, TRIM21, TCF7, RHD, RARB, TRBV20OR9-2, PTH, CDK5RAP3, VIM, POTEF, TWIST1, VEGFA, CCDC6, RAB11A, PRC1, DMTF1, LINC01194, HAVCR2, HUWE1, POSTN, RASSF1, HARS2, SLC25A19, MBD2, ABL1, PIK3C3, E2F1, HARS1, GC, FOXM1, FGF1, FCN1, FCGR3B, FCGR2A, DMP1, NOTCH1, CRH, CLU, CDKN2B, RUNX1, BCR, PARP1, ACTB, HLA-DQA1, HLA-DQA2, HLA-DQB1, AGFG1, MYC, MUC1, MTTP, MNT, MEFV, ABO, CXCL10, IL2RG, IL1RN, IL1B, IGHG3, IGFBP2, IFNG, IDH1, ICAM1, H3P10
-
Abusive Head Trauma
Wikipedia
Furthermore, shaking cervical spine injury can occur at much lower levels of head velocity and acceleration than those reported for SBS." [19] Other authors were critical of the mathematical analysis by Bandak, citing concerns about the calculations the author used concluding "In light of the numerical errors in Bandak’s neck force estimations, we question the resolute tenor of Bandak’s conclusions that neck injuries would occur in all shaking events." [20] Other authors critical of the model proposed by Bandak concluding "the mechanical analogue proposed in the paper may not be entirely appropriate when used to model the motion of the head and neck of infants when a baby is shaken." [21] Bandak responded to the criticism in a letter to the editor published in Forensic Science International in February 2006. [22] Diagnosis [ edit ] Diagnosis can be difficult as symptoms may be nonspecific. [1] A CT scan of the head is typically recommended if a concern is present. [1] While retinal bleeding is common, it can also occur in other conditions. [1] It is unclear how useful subdural haematoma, retinal hemorrhages, and encephalopathy are alone at making the diagnosis. [23] A skull fracture from abusive head trauma in an infant 3D CT reconstruction showing a skull fracture in an infant 3D CT reconstruction showing a skull fracture in an infant Triad [ edit ] While the findings of AHT are complex and many, [24] they are often incorrectly referred to as a "triad" for legal proceedings; distilled down to retinal hemorrhages , subdural hematomas , and encephalopathy. [25] AHT may be misdiagnosed, underdiagnosed, and overdiagnosed, [26] and caregivers may lie or be unaware of the mechanism of injury. [11] Commonly, there are no externally visible signs of the condition. [11] Examination by an experienced ophthalmologist is often critical in diagnosing shaken baby syndrome, as particular forms of ocular bleeding are quite characteristic. [27] Magnetic resonance imaging may also depict retinal hemorrhaging; [28] this may occasionally be useful if an ophthalmologist examination is delayed or unavailable. [ citation needed ] Conditions that are often excluded by clinicians include hydrocephalus , sudden infant death syndrome (SIDS), seizure disorders , and infectious or congenital diseases like meningitis and metabolic disorders . [29] [30] CT scanning and magnetic resonance imaging are used to diagnose the condition. [11] Conditions that may accompany AHT include bone fractures , injury to the cervical spine (in the neck), retinal bleeding, cerebral bleed or atrophy , hydrocephalus , and papilledema (swelling of the optic disc ). [12] The terms non-accidental head injury or inflicted traumatic brain injury have been suggested instead of Abusive head trauma or "SBS". [31] Classification [ edit ] The term abusive head trauma is preferred as it better represents the broader potential causes. [13] The US Centers for Disease Control and Prevention identifies SBS as "an injury to the skull or intracranial contents of an infant or young child (< 5 years of age) due to inflicted blunt impact and/or violent shaking". [32] In 2009, the American Academy of Pediatrics recommended the use of the term abusive head trauma to replace SBS, in part to differentiate injuries arising solely from shaking and injuries arising from shaking as well as trauma to the head. [33] SBS was previously believed to present with constellation of findings (often referred to as a " triad "): subdural hematoma ; retinal bleeding ; and brain swelling or encephalopathy – which has controversially been used to infer child abuse caused by violent shaking or traumatic shaking. [13] The diagnostic accuracy of the triad, linked to episodes of traumatic shaking is controversial with a 2016 systematic review finding limited scientific evidence associating the triad to episodes of traumatic shaking, and insufficient evidence for using the triad to identify such episodes. [13] The connection is controversial in part following cases where parents of children exhibiting the triad have, in addition to losing custody, been jailed or sentenced to death. [34] The Crown Prosecution Service for England and Wales recommended in 2011 that the term shaken baby syndrome be avoided and the term non accidental head injury ( NAHI ) be used instead. [35] Differential diagnosis [ edit ] Vitamin C deficiency [ edit ] Some authors have suggested that certain cases of suspected shaken baby syndrome may result from vitamin C deficiency. [36] [37] [38] This contested hypothesis is based upon a speculated marginal, near scorbutic condition or lack of essential nutrient(s) repletion and a potential elevated histamine level.
-
Hiv-Associated Neurocognitive Disorder
Wikipedia
Neuroimaging studies of HIV patients indicate that significant volume reductions are apparent in the frontal white matter, whereas subcortically, hypertrophy is apparent in the basal ganglia, especially the putamen . [16] Moreover, the results of some studies suggest loss of brain volume in cortical and subcortical regions even in asymptomatic HIV patients and patients who were on stable treatment. [24] A recent longitudinal study of a small representative cohort of HIV-positive patients on stable medication regiments suggests that this cortical atrophy is progressive, and is in part related to nadir CD4. [25] Cerebral brain volume is associated with factors related to duration of the disease and CD4 nadir; patients with a longer history of chronic HIV and higher CD4 nadir loss present with greater cerebral atrophy. [24] CD4 lymphocyte counts have also been related to greater rates of brain tissue loss. [26] Current factors, such as plasma HIV RNA, have been found to be associated with brain volumes as well, especially with regards to basal ganglia volume [24] and total white matter. [27] Loss of cortical grey matter oligodendrocytes might also contribute to the symptomatology. [28] Changes in the brain may be ongoing but asymptomatic, that is with minimal interference in functioning, making it difficult to diagnose HIV-associated neurocognitive disorders in the early stages. [29] Diagnostic criteria [ edit ] Marked acquired impairment of at least two ability domains of cognitive function (e.g. memory, attention): typically, the impairment is in multiple domains, especially in learning, information processing and concentration/attention.
-
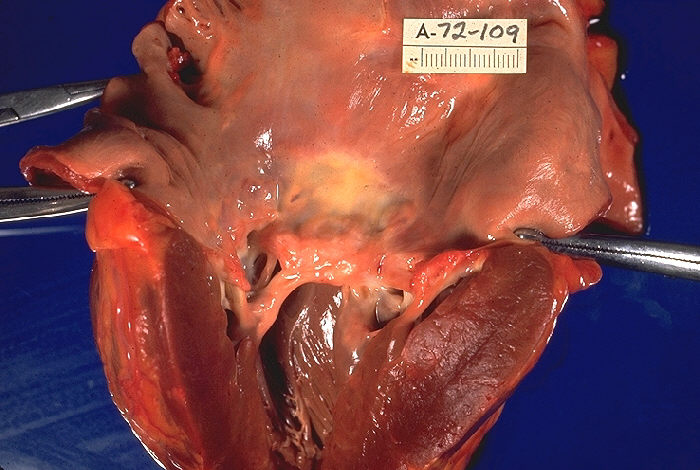
Tetralogy Of Fallot
Wikipedia
It is associated with chromosome 22 deletions and DiGeorge syndrome . [22] : 62 Specific genetic associations include: JAG1 , [23] NKX2-5 , [24] ZFPM2 , [25] VEGF , [26] NOTCH1 , TBX1 , and FLT4 . [27] Embryology studies show that it is a result of anterior malalignment of the aorticopulmonary septum , resulting in the clinical combination of a VSD, pulmonary stenosis, and an overriding aorta. [18] : 200 Right ventricular hypertrophy develops progressively from resistance to blood flow through the right ventricular outflow tract. [9] Pathophysiology [ edit ] Play media Video explanation The main anatomic defect in TOF is the anterior deviation of the pulmonary outflow septum. [9] This defect results in narrowing of the right ventricular outflow tract (RVOT), override of the aorta, and a ventricular septal defect (VSD). [28] Four malformations [ edit ] " Tetralogy " denotes four parts, here implying the syndrome's four anatomic defects. [2] This is not to be confused with the similarly named teratology , a field of medicine concerned with abnormal development and congenital malformations (including tetralogy of Fallot). ... Archived from the original on 2008-10-16 . Retrieved 2009-02-28 . ^ " ' Little Darth Vader' reveals face behind the Force" . 7 February 2011. ^ "Aide to All the Times Jimmy Kimmel Has Gotten Political" .ZFPM2, NKX2-5, JAG1, GATA4, TBX1, GATA6, GDF1, GJA5, CITED2, FLT4, GATA5, NKX2-6, FOXC2, FOXH1, HAND2, FGF8, FOXC1, HEY2, MKS1, BMP10, NTF3, INVS, DNAH5, PHC1, DOCK1, NRP1, HIRA, UFD1, NOTCH1, COMT, PTPN11, FN1, TAB2, MKKS, RFC2, RET, NOTCH2, NODAL, UBE2T, INTU, ROR2, CCDC22, SPECC1L, RAD51C, RAD51, DLL1, PTCH1, PIGN, NIPBL, RPH3A, TBL2, PAH, HIBCH, ATXN2, CERS1, RPL5, TDGF1, GTF2IRD1, PIGL, NR2F2, TGIF1, TPM1, UBE2A, SLC35A2, BAZ1B, CLIP2, WT1, XRCC2, ZIC2, CXCR4, ALX1, SEC24C, TTC37, SEMA3E, SIX3, RREB1, MAD2L2, SF3B4, SALL1, CDON, SHH, SKI, TMEM94, SKIV2L, PQBP1, SH2B3, RBM8A, WASHC5, FIG4, NID1, LINC02676, DACT1, STRA6, DDX59, NAA25, EHMT1, PALB2, CRELD1, NXN, ELN, SLX4, EPHB4, ERCC4, PRDM16, FANCM, FANCA, COX3, BRIP1, DISP1, FANCE, BRAF, ZFPM2-AS1, RNU4ATAC, ALX3, ARVCF, RERE, BRCA1, BRCA2, COL2A1, EOGT, MYRF, HECTD4, JMJD1C, CHD4, CHRM3, FANCD2, FANCC, FANCB, SALL4, COX2, COX1, SUFU, RAB23, DLL4, KCNAB2, FANCL, RFWD3, FANCI, CHD7, LIMK1, VAC14, FANCF, HDAC8, RBPJ, RBM10, FOXF1, FANCG, GTF2I, ARID1B, DOCK6, GP1BB, GLI2, ARHGAP31, FGFR1, GPC5, GAS1, GABRD, VEGFA, TBX20, CHDH, HAND1, GJA1, KDR, TGFB1, VANGL2, MIR421, MIR1233-1, SULT1E1, MTHFR, APOE, HIF1A, REN, PVR, PITX2, MMP9, NPPB, MEIS2, FBN2, MAVS, GPR42, HAS2, MMP3, HOXA1, ACKR3, DGCR8, MMP2, HTC2, JARID2, FBN1, LPA, LRPAP1, LSAMP, MAP4, MBNL1, FEZF2, SLC50A1, IQGAP1, F5, TSPYL2, ADRA2B, AGT, ALDH2, MIR625, MIR424, BRS3, CAD, CD48, CRKL, CRP, CYBA, ACE, DMD, FSD1L, DNAH8, DNMT1, DNMT3B, DNTT, CORO7, FSD1, DVL2, EDNRA, EGF, F2, MYL4, CHMP5, TCF21, SLN, SNAI2, SCO2, REC8, SMN1, SMN2, SSTR4, ADRA1A, TADA2A, TBX5, RGS6, WNT11, TFAP2B, CHD1L, TFAP2C, TTN, VIM, LPAR2, BEST1, MBD2, AVSD1, ALDH1A2, MBNL2, ACSM3, RYR2, RXRA, NTRK3, OPRD1, PBX1, PBX3, PDGFA, LAMP3, PECAM1, AATF, PFKL, SUN2, PLN, PTH, DICER1, RASA1, MMRN1, ROBO1, BVES, WWP2, PPARGC1A, ROCK1, CXCR6, ACTC1
-
Pain Management In Children
Wikipedia
Breastmilk or 'sugar' water has a similar effect, though studies in preterm infants have yet to be done. [26] Skin-to-skin care ( kangaroo care ) is thought be effective for pain control during painful procedures. [27] For children and adolescents who experience chronic pain- behavioral treatment, relaxation training, cognitive behavioral therapy (CBT) and acupuncture have been proven to be effective for some patients. [28] For recurrent abdominal pain a 2017 Cochrane review found some evidence that CBT and hypnotherapy were effective in reducing pain for the short term. [29] Medication [ edit ] Further information: Analgesic adjuvant Acute pain, chronic pain, neuropathic pain and recurrent pain in children is most often managed with medication.
-
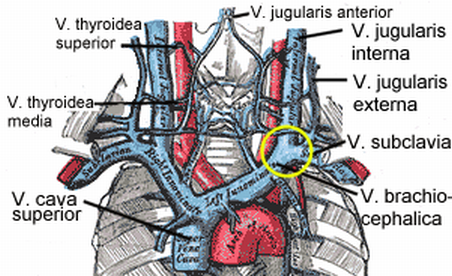
Nephritic Syndrome
Wikipedia
Jugular venous distention (JVD) may also be appreciated when visualizing the veins of the neck on physical exam. [9] Elevated blood pressure - Measured at least two separate times with at least two minutes between measurements using a sphygmomanometer or equivalent method. [28] Abnormal heart sounds - If the underlying cause is cardiac in nature (such as infective endocarditis ), then you may appreciate abnormal heart sounds during auscultation of the heart. [29] Laboratory testing [ edit ] If the physician is suspicious of a possible nephritic syndrome, then he/she may order some common lab tests including: Serum electrolytes - The kidney is one of the main regulators of electrolytes in the human body and measuring the different electrolyte levels using either a basic metabolic panel (BMP) or comprehensive metabolic panel (CMP) can be a useful indicator of the underlying pathology. [30] Serum creatinine - Also measured using a BMP or CMP , creatinine is one of the most important indicators of current kidney function and is used to calculate the glomerular filtration rate (GFR).
-
Viral Hemorrhagic Septicemia
Wikipedia
Clair . [27] The isolate was named MI03GL and was sequenced for its entire genome. [28] The North American genotype of the virus, in addition to moderate mortality to salmonid species, including various varieties of trout, is also proving virulent among a wide variety of warm-water species previously considered resistant to VHS.
-
Multiple Myeloma
Wikipedia
HIV/AIDS , organ transplantation , or a chronic inflammatory condition such as rheumatoid arthritis . [28] EBV-positive multiple myeloma is classified by the World Health Organization (2016) as one form of the Epstein-Barr virus-associated lymphoproliferative diseases and termed Epstein-Barr virus-associated plasma cell myeloma . ... Epidemiology [ edit ] Deaths from lymphomas and multiple myeloma per million persons in 2012 0-13 14-18 19-22 23-28 29-34 35-42 43-57 58-88 89-121 122-184 Age-standardized death from lymphomas and multiple myeloma per 100,000 inhabitants in 2004. [94] no data less than 1.8 1.8–3.6 3.6–5.4 5.4–7.2 7.2–9 9–10.8 10.8–12.6 12.6–14.4 14.4–16.2 16.2–18 18–19.8 more than 19.8 Globally, multiple myeloma affected 488,000 people and resulted in 101,100 deaths in 2015. [8] [9] This is up from 49,000 in 1990. [95] United States [ edit ] In the United States in 2016, an estimated 30,330 new cases and 12,650 deaths were reported. [7] These numbers are based on assumptions made using data from 2011, which estimated the number of people affected as 83,367 people, the number of new cases as 6.1 per 100,000 people per year, and the mortality as 3.4 per 100,000 people per year. ... National Cancer Institute . 1980-01-01 . Retrieved 28 November 2017 . ^ Ferri, Fred F. (2013). ... OCLC 888026338 . ^ Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, Dispenzieri A, Kumar S, Clark RJ, Baris D, Hoover R, Rajkumar SV (28 May 2009). "Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study" .FGFR3, CCND1, CRBN, IRF4, KRAS, NRAS, BRAF, CDKN2A, TNFRSF13B, CBX7, LIG4, BCL2, MYC, MUC1, MCL1, CSF3, IL6, ULK4, PSORS1C2, PARP1, MIR19A, CCL2, BCL2L1, PTHLH, NQO1, GSTT1, CDKN2C, CD86, HSPB1, EPHX1, BNIP3, HLA-A, SPARC, SOD2, CSF2, PRAME, BIRC3, KDM6A, NUAK1, IFNA2, MIR92A1, MAF, BTG1, IKZF1, TP53, B2M, PIK3CA, KMT2D, CDK4, PVT1, PCAT1, POU5F1, ELL2, HRAS, FOPNL, PTPN11, LPP, IDH2, PREX1, IDH1, CDCA7L, KLF2, GBA, CDH12, MAPK1, RPL36P20, NR3C1, LRRC31, LRRC34, CAMKMT, ACOXL-AS1, PIK3CG, PIK3CD, HGF, PIK3CB, PTEN, MTOR, ABCB1, PTPRC, MIR15A, FN1, CD44, CD40, CD38, CD34, MIR21, GRAMD1B, EZH2, MIR29B1, MIR29B2, CCL3, SDC1, DTNB, CDKN2B, MALAT1, IL1B, RNF165, IFNG, MIR4435-2HG, IGF1, IGH, CIB3, IL10, CTAG1B, MS4A1, PSORS1C1, CTNNB1, KIT, JAK2, CEP120, GGNBP1, MDM2, DNER, MET, BMF, NCAM1, NCK1, CKS1B, LCOR, ANKRD27, DIO1, TNFRSF11B, HSP90AA1, LRRIQ4, HLA-B, HIF1A, ATN1, CXCL12, H3P10, CD19, TNFSF11, PPT2, MSC, MYOM2, CACNG4, TNFSF10, ATP6V0E1P2, DNAH11, SOCS1, MYNN, VEGFA, CD274, NCAPH2, CDR3, BAK1, CXCR4, TNFRSF17, XBP1, PPT2-EGFL8, ABCG2, ANXA5, SMUG1, ATG5, HPSE, SRCAP, CHEK2, TNFSF13B, SP140, IKZF3, KLRK1, TRIM13, DKK1, AKT1, TOM1, ALB, HDAC6, TBC1D9, MED24, HDAC9, CASC19, NSD2, SLAMF7, BTK, ACOXL, USP8P1, TNF, CALM1, WAC, SP3, CALM2, CALM3, KLRC4-KLRK1, KRIT1, CCHCR1, RFWD3, KRT20, CASR, CASP3, SMARCD3, STAT3, MAPK3, SPG7, ARR3, TNFSF13, IL6R, CTAG1A, PRKAR1A, FAS, NR1I3, RTEL1, CXADR, CXCL8, MAPK8, TRAF3, ERN1, FGF2, SOST, CXADRP1, XPO1, XBP1P1, IL6ST, MIR34A, SUB1, NFKB1, MAFB, MIR221, PCNA, RB1, ABCC1, ANOS1, BSG, CCND2, IL17A, BCL6, MKKS, MIR20A, LOC102724971, EPO, ICAM1, MRPL28, LRPPRC, EIF4E, PTK2B, EPHB2, AICDA, SPP1, H3P9, VCAM1, IFNA1, MTHFR, IFNA13, NXT1, MAPK14, STOML2, CDK6, COL11A2, LOC102723407, IL4, PAX5, PRDM1, NM, PWWP3A, CD200, HSP90B1, PDCD1, MME, SOAT1, NOTCH1, AHSA1, OSM, AIMP2, MIR155, ANP32B, RBM45, VDR, PTGS2, BRD4, TENT5C, GRAP2, HSPA5, RNF19A, HMMR, TP73, IL2, IL3, INA, WWOX, HAS1, DIS3, POLDIP2, PIM2, MIR17HG, CCND3, ANXA2, CRK, BCR, CD28, BMI1, CD80, CD9, ACTB, APEX1, CASP8, CTLA4, CD81, CDKN1B, RUNX2, IL15, IL16, FRZB, GSTP1, CD47, CUX1, SCT, IL11, CD40LG, GGPS1, CTRL, CRP, NEDD8, SELPLG, GPRC5D, IGF1R, IGHV3-69-1, MIP, SPA17, CFLAR, MMP2, NEAT1, TGFB1, ANGPTL1, GDE1, LDLRAP1, CCDC54, CCR1, LRP1, MYD88, SLC7A5, ITGB1, KDR, COMMD3-BMI1, SGSM3, BMP2, MIR17, DCLRE1C, PTPN6, MAP2K7, ZHX2, DEPTOR, EGFR, EIF4G1, MIR19B1, FOXP3, MIR203A, JAG1, CD226, TNFRSF11A, JAK1, VCAN, NEK2, IL32, MIR214, IL1RL1, CD79B, SLC5A5, TICAM2, TIMP2, MIR210, PIM1, IGHV3OR16-7, NR2F2, ITGA4, DCTN6, H3P23, SMR3B, MARCKSL1, PTP4A3, HDAC3, B3GAT1, IFI27, JAG2, MMP9, BCL10, MSH3, HSPA4, NR0B2, HSF1, IGL, MIR135B, PSMD9, TNFRSF13C, NFKBIA, NFKB2, CDH1, TERT, SET, ARHGAP24, FCRL5, GFI1, GEM, MIR137, MVP, MIR16-1, PITX2, PF4, KLRC1, FLT4, EGF, ACTR1A, CDKN1A, ARFRP1, MIR152, SPRY2, MBL2, FRA16D, TMED7, GLI1, PMAIP1, GNA12, PPARG, CASP9, APAF1, MDM4, LGALS1, ZNRD2, TRAF6, DAP, RPSA, LINC01194, APOBEC2, GRN, SART3, VCP, WLS, UCA1, TMED7-TICAM2, ABL1, COX8A, BMP6, TFRC, LY9, CR2, TLR4, CXCR5, TRIAP1, MICA, JUN, TRPV2, TXN, UBE2I, CD82, TYMS, ITGA5, PARP14, PTPRA, PCDH10, PRKN, HAMP, PDK1, IL21, RAF1, CD276, CDK9, SPANXA2, RAC1, PVR, CD79A, KLF6, CDK2, PTGS1, POU2AF1, POU2F2, EPHA3, PTCH1, SRGN, RTL10, PSMD2, CDK1, MUL1, RPS6KA3, PAEP, S100A9, CEBPA, RAB8A, COL1A1, AURKA, TUG1, ABCC2, CCR7, MEG3, CD24, CCR5, COX1, MTR, TRIM63, SLC2A4, SLAMF1, SKP2, PMEL, NGF, ASCC2, SPHK2, CD27, CHEK1, OAS3, ODC1, ITGB7, RELN, AREG, HLA-G, FOS, SIRT1, HMOX1, CCN1, FLT1, IGHV3-52, FOXM1, TCL1A, IL1A, ATM, HLA-DRB1, CYP2C8, CFDP1, MIR192, HDAC4, TPX2, NAMPT, HNF4A, CXCR3, AGO2, USP14, HP, CRTC1, PKD2L1, TNFRSF10A, DAZAP2, ALDH1A1, MIR125A, FOXP1, GJA1, SND1, XIAP, HLA-E, CIB1, XRCC4, RASSF1, E2F1, ADM, GEMIN4, GDF15, PRDX5, XRCC5, BCL9, CTAG2, CXCL10, CBLL2, BDNF, MIR497, BGLAP, EPAS1, ZDHHC9, ANGPT1, ESR1, EIF4G2, SPANXA1, HK2, USP7, USP5, EZH1, ADAR, F9, DPP4, RRM1, MELK, MIB1, TCL1B, ALPP, ALPI, CCL20, CX3CL1, APC, ADAMTS9, CCL5, RUNX2-AS1, SAT1, ACKR3, DICER1, EEF1E1, CCL4, CD33, ING4, RET, ADCYAP1, CD70, PTK2, CD74, SEMA3A, DLC1, CARM1, ADCYAP1R1, CASC3, PAK4, TAS2R38, SAMSN1, PLK2, PPP1R13L, CD3EAP, ABO, PPARGC1A, CCR2, NR0B1, KLHL1, AKT2, RELA, CIP2A, LOC105379528, MAGEC1, ALCAM, RAP1A, BCL2L11, RAN, MIR767, ABCB6, KDM1A, RAD51, DIABLO, RAD50, ZNF197, AHR, RETN, SLC22A2, ADIPOQ, H3P12, TAM, TIMP1, USP9X, THPO, THBS1, ATF4, SETD2, MKNK1, IL17D, TNFRSF10D, PSMG1, TRBV20OR9-2, BECN1, MBTPS1, HNF1A, HPGDS, TNFSF14, TAZ, MPC1, ATR, IL23A, BAX, BIK, XRCC1, F11R, MZB1, IGK, ATRAID, IL22, IL21R, TXNRD1, UCHL5, SLC40A1, TSC1, MAGEC2, BST2, MAFK, CCDC6, DCDC2, DLK1, XAF1, SFRP2, VHL, MYEOV, CD6, BIRC5, IL17RB, SMPD1, AURKB, SETBP1, NOG, SLC16A1, MAP3K7, SLC7A4, MAPKAPK2, KDM3A, BIRC2, CD14, TRIP13, CD163, MYDGF, SYCE1L, SOCS3, CCNE1, SP1, SYK, TNFRSF10B, ANKHD1, STIM1, CASP10, IQGAP1, STAT1, MIR1271, SSX2, FASLG, LAMTOR1, SST, SQSTM1, CCNA1, RUNX1, NBEA, HSPB3, EPHA4, PTH, IGHG3, CSF1R, HSPB2, MIR324, CEACAM6, GCLC, MTRR, SCARNA22, GLS, MRE11, ZFP82, JUNB, MIR106B, MIRLET7E, JUND, GRK6, CRYZ, MPO, MIR144, ISG20, IPP, NFE2L2, RCBTB2, NPM1, G6PD, MIR148A, MIR146A, NOTCH2, GABPA, IAPP, GAPDH, MIR145, LYST, SSX2B, GAS6, HTC2, GATA1, CCR6, AGFG1, YBX1, DUSP2, PRRT2, LDHA, LEP, HDAC1, AZIN2, LGALS3, LGALS9, HNRNPA1, MMP13, MIR99B, TPPP2, DNMT1, LTA, HMGB1, LPL, LIG3, POU5F1P4, EGLN3, MEFV, MTDH, PRIMA1, GSTM1, MMP7, KMT2A, GADL1, WNT3A, TNPO1, MICB, CD99, LOC390714, HNRNPA1P10, LAG3, MGMT, LAIR1, KITLG, IRF8, ING1, EDNRA, PLG, MIR22, CYP2C19, DCC, CDK5, NCR3, FCGR2A, MAGT1, FCGR3A, PDPK1, FCGR3B, LBH, EIF4A2, CYP1A1, IFNAR1, MIR202, IL27, PLK1, EGR1, EXT1, EP300, IGHA1, EPS8, MIR29A, MIR451A, PRKCB, PRKCA, IKBKB, PRKAB1, IL1RN, MIR223, IL2RA, PPP1R1A, CDH2, IGFBP2, TRIM56, POU5F1P3, CDKN3, MIR375, FOSB, ROR2, CFL1, P2RX7, P4HB, TNFRSF9, MIR342, FOLH1, ASXL1, SERPINA5, MIR18A, PRDX1, DDIT3, MIR186, FLT3, FGFR1, CD52, CX3CR1, CRTC2, MIR363, RABGEF1, MCAT, ST3GAL6-AS1, DARS-AS1, MIR340, KCNRG, SHISA8, STAC3, MCIDAS, PABPC1P1, BLOC1S2, PPP1R16B, MIR338, ZNF763, SPDYA, IGHV5-78, LINC01193, STEAP1, GPSM1, AATF, FOXD3, HOXC-AS3, PTENP1-AS, NUPR1, PABPC1, ABCB5, MYCBP, MIR335, RICTOR, LAMA5-AS1, BBC3, MIR331, NANOS3, CLEC4D, MIR425, TINF2, LINC00515, DIMT1, FBXO9, PDCD4, PHF19, GSTK1, APEX2, MMP28, IBTK, MRAS, MIR197, MLXIP, MIR193A, CLSTN1, LINC02605, CARD8, RNA28S5, MIR182, RNA28SN4, MMRN1, ATF6, RPIA, MIR30E, PRAL, MIR320A, MIR150, MTCO2P12, TNIK, SMG1, MTF2, MIR30D, CIC, VSIG4, MIR301A, RPP14, CIT, CDC37, GLMN, MIR28, MIR27A, MIR26B, MIR23B, MIR30A, SLC2A6, MIR30C1, PTENP1, PADI2, H3P8, MIR30C2, CHP1, H3P13, MIR215, KDM6B, MIR34B, MAST4, MAPK8IP2, PSD4, LPAR3, CSAG3, PADI4, CSAG2, MIR93, LIN28B, DAPK2, NUP62, ANXA2R, SGK3, HES5, PSMA3-AS1, MAFF, MIR9-3, CA13, RCHY1, POT1, SOSTDC1, SEZ6L, MIR9-2, RN7SL263P, IGHV4-34, ACSBG1, MIR130A, RHOBTB2, MIR127, DNMBP, USP24, MIR126, ZNF629, PPP1R13B, MIR124-1, MIR10A, CERNA3, SIRT3, MIRLET7B, RYBP, MIR9-1, SF3B1, PERCC1, IRAIN, CABIN1, TRAT1, ZUP1, FERMT3, PAG1, USE1, PBK, GPRC5C, PROK1, SLC2A9, RPAIN, TCHP, MAGED4, RNASEH2C, TCIM, PMEPA1, VTRNA2-1, CHPT1, SLC12A9, KREMEN1, EIF2A, CIAPIN1, RASSF4, CRISPLD2, PDXP, DOT1L, NUP133, NLRP2, ORAI1, MIR1246, PRINS, SNORD14B, LGR4, RBPMS, ZNF331, CAMK2N1, ATAD1, HES6, HAVCR2, MIR631, RIOX2, SOX6, SUPT20H, MIR33B, SPZ1, SNORD35B, DEPDC1, MIR532, MIR610, PCBP4, PLSCR4, DLL1, GOPC, ZKSCAN3, ASRGL1, UBE2O, ZC3H12A, DHDDS, TXNDC15, LIN28A, DHX40, TINAGL1, IFIH1, FAM72D, MMP25, GORASP1, PDIA2, OIP5-AS1, RAPH1, NEIL1, WNK1, SLC2A11, TRPM8, MAPKAP1, CEP70, ULBP2, PDCD1LG2, WDR19, RALGAPB, TP73-AS1, RNA28SN5, MIR765, RPTOR, SESN2, SEMA6A, RASSF5, KIF18A, POTEF, HPSE2, TLR10, CTDSP1, RHOU, MAGED4B, SLC38A1, SCPEP1, ZNF436, BACH2, COL18A1, SNORD14C, MSTO1, SNORD14D, SNORD14E, MIR193B, TENT5D, DERL1, SLC30A8, BHLHA15, CLEC12A, FEZF1-AS1, SLC2A12, MIR520G, DTX3L, GOLT1B, IL23R, ANGPTL4, ZNF569, SLC45A2, IL34, LINC00328, ARMH4, DCTN4, MIR509-1, SLC25A37, RHOV, MIR4741, PDIA3P1, MPEG1, LAMTOR2, MCTS1, ALKBH3, UBE2T, MARCHF8, MYLIP, MIR410, DEL13Q14, RMC1, PSC, MIR4505, PRSS55, PADI1, MIR485, PIKFYVE, SLC2A8, MIR490, ERO1A, MIR4449, SOX8, RPL17-C18orf32, A2ML1, MS4A4A, EGLN1, MIR155HG, NLRP3, SLC35A4, TP53RK, SHC3, TERF2IP, CYTOR, DDX4, DDIT4, FOXP2, ZNF501, FBLIM1, PPP1R12C, TET2, DPP8, DCAF15, CASZ1, NSD3, SLC25A21, ITPRIP, ADA2, MIR4254, FZR1, LINC01672, CACUL1, MIB2, NRSN1, SCARA3, PASD1, SLCO6A1, EVL, CPT1C, LINC01234, ETV7, LINC00461, SIRT6, SEZ6, SNHG18, NBAS, SF3B6, RASD1, CRNDE, ZNF44, IGLL5, RITA1, NAT1, GLIPR1, IGF2, IGHM, IGHG1, IGHDOR15@, IGFBP7, IGFBP4, IGF2R, IFNB1, IL2RB, IDUA, ID1, HSPA8, HSPA1B, HSPA1A, HES1, JCHAIN, IL2RG, ITGAV, ILF2, IRF1, IRAK1, INPPL1, INPP5D, IDO1, ILK, IL18, IL5, IL13, IL12RB2, IL11RA, IL10RB, IL10RA, IL7, HOXC10, HOXB7, HOXA11, GFER, GPR42, CCR10, SFN, GOT2, GLO1, GLB1, GDF1, HOXA1, GCG, GBP1, GARS1, GAB1, XRCC6, IFI6, GRB2, GRM3, GTF2H1, GZMB, H2AX, HAS2, HCK, HDAC2, HFE, HGFAC, HIC1, HLA-DOB, HLA-DQB1, HLA-DRB5, HMBS, NR4A1, HOXA@, ITGAL, ITGAX, MMP24, COX2, NEDD4, NDP, NBN, NAGLU, MUC4, NUDT1, MSX1, NOS2, MRC1, MPZ, MPST, MPG, MMP15, MMP8, NOS1, NOS3, ITGB2, PAFAH1B1, PDC, PCYT1A, PCM1, PBX3, PAM, SERPINE1, FURIN, NOTCH3, PEBP1, ORC4, ROR1, NTF3, NPPB, NPC1, MMP1, MAP3K10, MLH1, STMN1, CD180, LTBP3, LIF, LGALS8, LEPR, LCN2, LAMP1, MITF, LAMA5, KRT81, KPNA2, KLRB1, KIR3DS1, ITGB3, EPCAM, MARCKS, SMAD1, SMAD2, SMAD3, SMAD5, MAGEA1, MAOA, MAX, MBD1, CD46, MEIS2, MAP3K5, MFAP1, MGST1, CIITA, MIF, FUT1, FOLR2, FOLR1, CASP2, CD247, CD1D, CCNB1, CCK, RUNX3, CAV1, CANX, TNFRSF8, CALCR, DDR1, BUB1B, BTF3P11, KLF9, BRS3, CD22, TNFSF8, FLII, CHUK, CNR2, CCR4, CMA1, TPP1, CLCN5, CISH, CHI3L1, CD48, CDKN2D, CDK7, CDC42, CDC25C, CDC20, CD68, BRCA2, BMPR1A, CEACAM1, AHCY, AMY1B, AMY1A, AMBP, ALOX5, ALK, CRYBG1, AGER, BCL3, ADRA2B, ADRA1A, ADK, ACVRL1, ACVR2A, ACP5, AMY1C, ANG, ANGPT2, ANXA1, APOA1, APOB, APRT, ARF1, ARG1, RHOH, ARNT, ARNTL, ATF3, ATP7A, AXL, BCHE, BCL2A1, COL4A1, SLC31A2, MAP3K8, EPOR, ETFA, ERG, ERCC3, ERCC2, ERBB3, ERBB2, ENO1, EDN1, ENDOG, EMX2, ELF4, ELAVL2, EIF4EBP1, EEF1A2, ETS1, F2, F3, F10, FASN, FCAR, FCER2, FCGR2B, FCGRT, FDPS, FGA, FGF1, FGF13, FGFR4, FH, FHIT, FLI1, EDNRB, LPAR1, CPOX, CYP1B1, DCN, DAPK1, CYP3A4, CYP2D6, CYP2C9, CYP2B6, CYLD, S1PR1, CTSK, CCN2, CSNK1A1, HAPLN1, CREBBP, CREB1, ACE, DDB1, DDX5, TIMM8A, DHCR24, DHFR, DIO3, DKC1, DLD, DMD, DNASE1, DNMT3A, SLC26A3, TSC22D3, DUSP5, DUSP8, TYMP, ENPP1, ENPP2, PDZK1, TNKS, TNFRSF10C, TNFRSF18, TNFRSF6B, ADAM23, TNFSF9, RIPK1, DYNLL1, PROM1, NCOA1, USO1, KMO, IKBKG, ITGA8, ENC1, SOCS2, DLEU2, UCN, CDY1, LPAR2, PDCD5, CBFA2T2, SLC16A3, SLC16A4, FCGR2C, PRC1, NR1I2, SEMA5A, MAP3K14, BTRC, MBD2, SPHK1, ARHGEF7, OFD1, SMARCA5, CUL4A, VWF, ZAP70, YWHAE, YY1, WT1, WNT5A, WEE1, VRK1, DYRK2, TRPV1, BEST1, VIM, VDAC1, USP1, NR1H2, ZNF70, TRIM25, ZMYM2, LAPTM5, EVI5, DEK, BAG6, RNF217-AS1, TFPI2, FGF23, CLLS2, NCOA3, USP9Y, ARID1A, TRRAP, BCAR3, RECK, SLC16A7, MTA2, PTTG1, CDK2AP2, CRTAP, TACC3, PRMT5, ST3GAL6, RACK1, STUB1, TRIB1, AKT3, PSMD14, PSME3, PDZK1IP1, ARPC5, EDIL3, ABCC5, SEMA4D, PRDX4, ARPC1A, CXCL13, GNLY, COLEC10, KHDRBS1, CXCR6, NRG3, OGA, NES, KDM5B, ZMYND11, CCL27, LILRB1, SLC26A1, MALT1, BCL2L10, SCO2, PDLIM7, NCR2, CHD1L, VPS4B, TBPL1, NAPSA, HOMER1, NCR1, COX5A, USP15, KL, LONP1, KLF4, ASIC3, CD83, ITGB1BP1, MINPP1, SOX13, APOBEC3B, RBM39, CYTIP, CARTPT, NCOR2, TTI1, HERPUD1, DCAF1, RAPGEF5, MTSS1, SPATA2, GAB2, TELO2, UBAP2L, DLEC1, UGCG, UCHL1, PFKFB3, RBBP8, RNASE2, RGS1, RENBP, REG1A, RECQL, OPN1LW, RBBP5, RNF6, RARA, RANBP2, RAG2, MOK, RAD23B, PTN, RNASE3, RNH1, UBE2G1, ATXN1, SELL, SELE, CXCL11, CCL8, CCL7, SCN7A, RYR1, SNORD15A, RRM2, RPS27A, RPS27, RPL17, RPL5, RPA1, PTBP1, PSME2, PSMD10, PLAUR, PPP3CA, PON1, POLD1, PMP22, PML, PLXNB1, PLAT, PSMD4, PKM, PIK3R2, PIK3R1, PIGA, ABCB4, PFN1, PRB1, PRKAA1, PRKAA2, MAPK7, MAPK9, MAP2K1, PSAP, PSMA3, PSMA6, PSMB1, PSMB4, PSMB5, PSMB8, PSMB9, PSMB10, PSMC1, PSMC6, SELP, SEMG1, SELENOW, TERF1, KLF10, THBS2, THBD, TGFA, TFPI, NAT2, TERC, ADAM17, TEAD1, TDG, TCF19, GCFC2, SERPINA7, TAPBP, TIMP3, TJP1, TLR3, TSPAN7, TMSB4X, TNFAIP3, TNFRSF1B, TOP1, TPO, CRISP2, TRAF2, TRPC1, TSC2, TSHR, TTK, TYRO3, UBA1, TAP2, SYT1, SFRP1, SKIV2L, SNCA, SIGLEC1, SMO, SMARCB1, SLC3A2, SKP1, SKI, SYCP1, SIM2, SH3GL3, SH3GL1, SGK1, SRSF2, SFRP5, FSCN1, SOD1, SORL1, SOX1, SOX2, SOX4, SOX11, SRI, ITPRID2, SSTR4, SSX1, SSX4, STAT5A, STAT5B, STK11, STX4, SULT1A1, TERF2
-
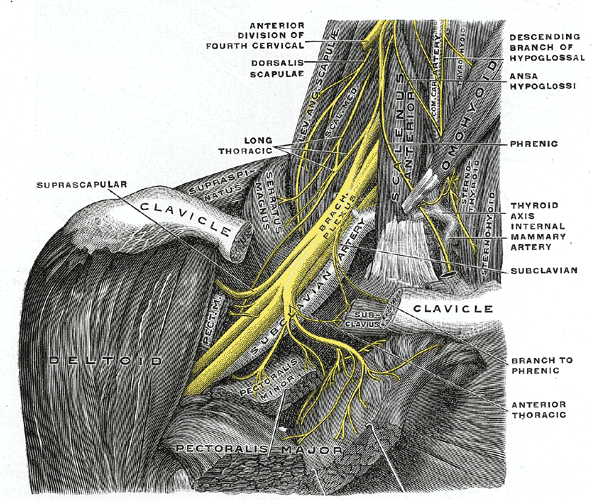
Brachial Plexus Injury
Wikipedia
Recovery takes place without wallerian degeneration . [26] [27] Axonotmesis : Involves axonal degeneration, with loss of the relative continuity of the axon and its covering of myelin, but preservation of the connective tissue framework of the nerve (the encapsulating tissue, the epineurium and perineurium , are preserved). [26] [28] Neurotmesis : The most severe form of nerve injury, in which the nerve is completely disrupted by contusion, traction or laceration.
-
First Red Scare
Wikipedia
The Senators heard various views of women in Russia, including claims that women were made the property of the state. [27] The press reveled in the investigation and the final report, referring to the Russians as "assassins and madmen," "human scum," "crime mad," and "beasts." [28] The occasional testimony by some who viewed the Bolshevik Revolution favorably lacked the punch of its critics.
-
Hiv/aids In New York City
Wikipedia
Joseph credited activists such as Yolanda Serrano , the head of the Association for Drug Abuse Prevention and Treatment (ADAPT) for calling attention to how the problem of addiction was far less dire than the overwhelming number of deaths from the rapidly spreading AIDS epidemic. [21] : 6 However, at the start of Mayor David Dinkins administration in 1990, the only legal needle exchange program was shut down. [25] Following new studies that demonstrated reduction in HIV infection rates, new programs restarted in New York City in 1992. [28] Comparison with San Francisco [ edit ] No two cities were more prominent in the battle against AIDS in the 1980s than New York and San Francisco. ... Rockwell; D.C.D. Jarlais; R. Elovich (28 December 2006). "Harm reduction theory: Users culture, micro-social indigenous harm reduction, and the self-organization and outside-organizing of users' groups" . ... Koch Collection, Koch Oral History Collection. Koch, Edward I. (28 July 2011). "LaGuardia and Wagner Archives Oral History: Mayor Ed Koch discusses the needle exchange program" (Video) .
-
Scurvy
Wikipedia
In the 1497 expedition of Vasco da Gama , the curative effects of citrus fruit were already known [25] [26] and confirmed by Pedro Álvares Cabral and his crew in 1507. [27] The Portuguese planted fruit trees and vegetables in Saint Helena , a stopping point for homebound voyages from Asia, and left their sick, who had scurvy and other ailments, to be taken home by the next ship if they recovered. [28] In 1500, one of the pilots of Cabral 's fleet bound for India noted that in Malindi , its king offered the expedition fresh supplies such as lambs, chickens, and ducks, along with lemons and oranges, due to which "some of our ill were cured of scurvy". [29] [30] Unfortunately, these travel accounts did not stop further maritime tragedies caused by scurvy, first because of the lack of communication between travelers and those responsible for their health, and because fruits and vegetables could not be kept for long on ships. [31] In 1536, the French explorer Jacques Cartier , exploring the St. ... Westmead Institute for Medical Research . 28 November 2016. Archived from the original on 23 August 2017 .
-
Chronic Cerebrospinal Venous Insufficiency Controversy
Wikipedia
Problems with the innominate vein and superior vena cava have also been reported to contribute to CCSVI. [26] A vascular component in MS had been cited previously. [27] [28] Several characteristics of venous diseases make it difficult to include MS in this group. [14] In its current form, CCSVI cannot explain some of the epidemiological findings in MS. ... Consensus document of the International Union of Phlebology (IUP)-2009". International Angiology . 28 (6): 434–51. PMID 20087280 . ^ "CCSVI – current studies" . mssociety.org.uk .
-
Doping In Sport
Wikipedia
The chairman of the Dutch cycling federation, Piet van Dijk, said of Rome that "dope – whole cartloads – [were] used in such royal quantities." [28] The 1950s British cycling professional Jock Andrews would joke: "You need never go off-course chasing the peloton in a big race – just follow the trail of empty syringes and dope wrappers." [29] The Dutch cycling team manager Kees Pellenaars told of a rider in his care: I took him along to a training camp in Spain.
-
Lung Cancer
Wikipedia
As more damage accumulates, the risk for cancer increases. [24] Smoking [ edit ] Tobacco smoking is by far the main contributor to lung cancer. [4] Cigarette smoke contains at least 73 known carcinogens , [25] including benzo[ a ]pyrene , [26] NNK , 1,3-butadiene , and a radioactive isotope of polonium – polonium-210 . [25] Across the developed world, 90% of lung cancer deaths in men and 70% of those in women during the year 2000 were attributed to smoking. [27] Smoking accounts for about 85% of lung cancer cases. [7] A 2014 review found that vaping may be a risk factor for lung cancer but less than that of cigarettes. [28] Passive smoking – the inhalation of smoke from another's smoking – is a cause of lung cancer in nonsmokers.