-
Back Pain
Wikipedia
In general, fatigue can worsen pain. [2] A few studies suggest that psychosocial factors such as on-the-job stress and dysfunctional family relationships may correlate more closely with back pain than structural abnormalities revealed in X-rays and other medical imaging scans. [23] [24] [25] [26] Diagnosis [ edit ] Diagnostic work-up for acute back pain. [27] Initial assessment of back pain consists of a history and physical examination. [28] Important characterizing features of the back pain include location, duration, severity, history of prior back pain, and possible trauma. Other important components of the patient history include age, physical trauma, prior history of cancer, fever, weight loss, urinary incontinence, progressive weakness, or expanding sensory changes, which can elicit red flags indicating a medically urgent condition. [28] Physical examination of the back should assess for posture and deformities. ... Retrieved April 30, 2018 . ^ a b author., Stern, Scott D. C. (2014-10-28). Symptom to diagnosis : an evidence-based guide .PLAT, HTR2A, HGD, HLA-B, LRP5, SUFU, CFI, TP53, SYNE2, PALLD, KRAS, SEC63, DLL3, LMNA, LMX1B, HELLPAR, CFH, CD46, TCIRG1, PLEKHM1, TMEM94, NF2, PDGFB, PRKCSH, SLC20A2, SMARCB1, SOX5, VCP, SMAD4, SYNE1, DIS3L2, BRCA1, FLI1, CAPN3, FHL1, CDKN2A, TMEM43, EMD, PALB2, VANGL1, CLCN7, ATP7B, MESP2, BRCA2, DCC, CRP, SMS, RSS, SPNS1, NARS1, CNTN3, TSPO, GLA, ZNF35, IS1, SPARC, AR, TP63, TNF, IL10, CXCL8, CD38, CSF1R, CD6, IL6, MOCOS, DEF6, MCF2L2, TRAPPC9, RGS6, BEST1, CREB3L1, GDF5, ZC4H2, BAS, SPTLC1, ACR, TNFRSF1A, CCN1, STS, AVP, CALCA, CD74, CSF1, CSF2, ACE, DMD, DPEP1, ESR1, FTH1, HLA-C, HMGB1, HP, IL1B, SPINK1, IL1RN, IL4, LAMC2, LEP, SMCP, MIF, AMH, MOS, MPO, NDUFS4, OGN, PIK3C2A, RASA1, CCL2, MMP1
-
Dihydrolipoamide Dehydrogenase Deficiency
Gene_reviews
Features of the Early-Onset Neurologic Phenotype View in own window Disease Features Frequency 1 % Clinical presentation 2 Hypotonia 16/25 64 Developmental delay 12/25 48 Emesis 12/25 48 Hepatomegaly 10/25 40 Lethargy 8/25 32 Seizures 7/25 28 Spasticity (hypertonia &/or hyperreflexia) 7/25 28 Leigh syndrome phenotype 6/25 24 Failure to thrive 6/25 24 Microcephaly 5/25 20 Vision impairment 4/25 16 Ataxia 3/25 12 Cardiac involvement 3/25 12 Laboratory abnormalities Metabolic acidosis 3 22/25 88 ↑ plasma lactate 4 18/25 72 ↑ urine α-ketoglutarate 4 13/25 52 Hypoglycemia 5 12/25 48 ↑ plasma BCAA 4 10/25 40 ↑ transaminases 11/25 44 ↑ urine branched-chain ketoacids 4 7/25 28 Hepatic failure 5/25 20 ↑ plasma allo-isoleucine 4 4/25 16 Low free plasma carnitine 6 3/25 12 Hyperammonemia 7 4/25 16 Includes only individuals biochemically confirmed to have DLD deficiency BCAA = branched-chain amino acids 1.
-
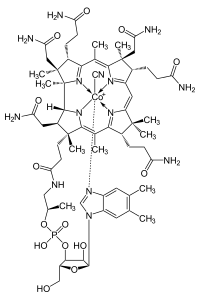
Vitamin B12 Deficiency
Wikipedia
B 12 deficiency is more common in the elderly because gastric intrinsic factor, necessary for absorption of the vitamin, is deficient, due to atrophic gastritis. [28] Impaired absorption of vitamin B 12 in the setting of a more generalized malabsorption or maldigestion syndrome. ... This process does not affect absorption of small amounts of B 12 in supplements such as multivitamins, since it is not bound to proteins, as is the B 12 in foods. [28] Surgical removal of the small bowel (for example in Crohn's disease ) such that the patient presents with short bowel syndrome and is unable to absorb vitamin B 12 . ... Retrieved 2013-12-31 . ^ "Test used to diagnose B 12 deficiency may be inadequate" . news-medical.net. October 28, 2004. Archived from the original on September 29, 2007 .
-
Binge Eating Disorder
Wikipedia
In 2013 it gained formal recognition as a psychiatric condition in the DSM-5. [26] The 2017 update to the American version of the ICD-10 includes BED under F50.81. [27] ICD-11 may contain a dedicated entry (6B62), defining BED as frequent, recurrent episodes of binge eating (once a week or more over a period of several months) which are not regularly followed by inappropriate compensatory behaviors aimed at preventing weight gain. [9] Diagnostic and Statistical Manual [ edit ] Previously considered a topic for further research exploration, binge eating disorder was included in the Diagnostic and Statistical Manual of Mental Disorders in 2013. [28] Until 2013, binge eating disorder was categorized as an Eating Disorder Not Otherwise Specified , an umbrella category for eating disorders that don't fall under the categories for anorexia nervosa or bulimia nervosa. ... However, the prevalence of eating disorders is increasing in other non-Western countries. [51] Though the research on binge eating disorders tends to be concentrated in North America, the disorder occurs across cultures. [52] In the USA, BED is present in 0.8% of male adults and 1.6% of female adults in a given year. [28] The prevalence of BED is lower in Nordic countries compared to Europe in a study that included Finland, Sweden, Norway, and Iceland. [51] The point prevalence ranged from 0.4 to 1.5 percent and the lifetime prevalence ranged from 0.7 to 5.8 percent for BED in women. [51] In a study that included Argentina, Brazil, Chile, Colombia, Mexico, and Venezuela, the point prevalence for BED was 3.53 percent. [53] Therefore, this particular study found that the prevalence for BED is higher in these Latin American countries compared to Western countries. [53] The prevalence of BED in Europe ranges from <1 to 4 percent. [54] Co-morbidities [ edit ] BED is co-morbid with diabetes, hypertension, previous stroke, and heart disease in some individuals. [46] In people who have obsessive-compulsive disorder or bipolar I or II disorders, BED lifetime prevalence was found to be higher. [46] Additionally, 30 to 40 percent of individuals seeking treatment for weight-loss can be diagnosed with binge eating disorder. [32] Underreporting in Men [ edit ] Eating disorders are oftentimes underreported in men. [51] Underreporting could be a result of measurement bias due to how eating disorders are defined. [51] The current definition for eating disorders focuses on thinness. [51] However, eating disorders in men tend to center on muscularity and would therefore warrant a need for a different measurement definition. [51] Further research should focus on including more men in samples since previous research has focused primarily on women. [51] History [ edit ] The disorder was first described in 1959 by psychiatrist and researcher Albert Stunkard as " night eating syndrome " (NES). [55] The term "binge eating" was coined to describe the same bingeing-type eating behavior but without the exclusive nocturnal component. [56] There is generally less research on binge eating disorder in comparison to anorexia nervosa and bulimia nervosa. [5] See also [ edit ] Prader–Willi syndrome References [ edit ] ^ Agüera, Zaida; Lozano-Madrid, María; Mallorquí-Bagué, Núria; Jiménez-Murcia, Susana; Menchón, José M.; Fernández-Aranda, Fernando (28 April 2020).
-
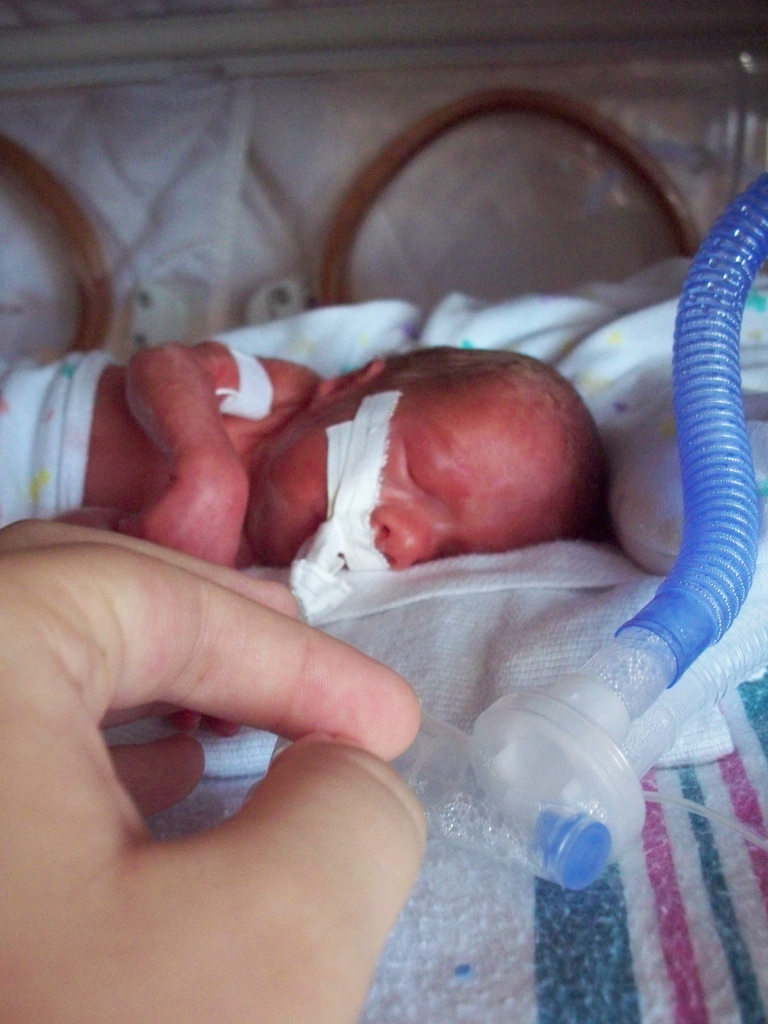
Neonatal Infection
Wikipedia
Risk factors for neonatal infection within the first week Factor Notes References prematurity birth before 40 weeks gestation [8] meconium aspiration inspiration of stool in utero [14] Postpartum endometritis inflammation of the uterus after the birth [14] low birth weight < 40 weeks gestation [8] [15] premature rupture of membranes <12 hours [5] [8] [15] [28] prolonged premature rupture of membranes >12 hours [5] [28] pre-term onset of labor labor begins before 40 weeks gestation [8] [15] chorioamnionitis inflammation of the fetal membranes(amnion and chorion) due to a bacterial infection [8] vaginal discharge abnormal discharge can be a result of an infection [8] tender uterus discomfort when the uterus is examined [29] rupture of membranes <12 hours [5] prolonged rupture of membranes >12 hours) [8] [29] in utero infection with pathogens the period of infection allows for the logarithmic growth of pathogens [7] maternal urinary tract infection bladder and/or kidney infection [8] prolonged labor [29] vaginal examinations during labor risk increases with the number of vaginal examinations during labor [8] [29] maternal colonization with group B streptococcus the presence of this bacterium is usually asymptomatic [5] [8] previous baby with early-onset GBS infection [8] [29] gender males are more susceptible;This risk declines after respiratory distress syndrome is treated [15] multiples risk is increased for the firstborn [15] iron supplementation iron is a growth factor for some bacteria [15] maternal intrapartum fever > 38 °C [8] [28] after insertion of intravenous line may introduce pathogens into the circulation [15] immature immune system [15] invasive medical procedures may introduce pathogens into the circulation [15] hypoxia unexpected resuscitation after birth [15] [29] low socioeconomic status [15] hypothermia relatively low blood temperature [15] metabolic acidosis a pH imbalance in the blood [15] obstetrical complications [15] prevalence of resistant bacteria in the neonatal intensive care unit nosomial populations [15] maternal exposure to Toxoplasmosis gondii a parasite present in cat litter and other animal excrement [27] Risk factors for late onset for neonatal infection >one week after birth Factor Notes References after insertion of an intravenous line hypothermia poor feeding lethargy more likely to develop osteoarthritis soft tissue infection meningitits [15] Mother resides in endemic malaria area [25] The risk for developing catheter-related infections is offset by the increased survival rate of premature infants that have early onset sepsis.
-
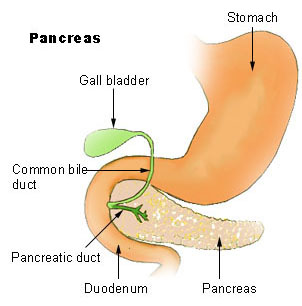
Johanson–blizzard Syndrome
Wikipedia
Variability of the phenotype, associated with residual ubiquitin ligase activity in some patients, has also been attributed to hypomorphic mutations occasionally found in either of the carrier parents. [1] [3] [14] [21] [22] The UBR1 gene is located on human chromosome 15 . [14] Diagnosis [ edit ] Exocrine [ edit ] The most prominent effect of Johanson–Blizzard syndrome is pancreatic exocrine insufficiency . [1] [23] [24] [14] [25] Varying degrees of decreased secretion of lipases , pancreatic juices such as trypsin , trypsinogen and others, as well as malabsorption of fats and disruptions of glucagon secretion and its response to hypoglycemia caused by insulin activity are major concerns when Johanson–Blizzard syndrome is diagnosed. [1] [3] [26] Associated with developmental errors, impaired apoptosis , and both prenatal and chronic inflammatory damage, necrosis and fibrosis of the pancreatic acini (clusters of pancreatic exocrine gland tissue, where secretion of pancreatic juice and related enzymes occurs), pancreatic exocrine insufficiency in Johanson–Blizzard syndrome can additionally stem from congenital replacement of the acini with fatty tissue . [1] [3] [26] [27] [28] Near total replacement of the entire pancreas with fatty tissue has also been reported. ... "The N-end rule pathway of protein degradation" . Genes Cells . 2 (1): 13–28. doi : 10.1046/j.1365-2443.1997.1020301.x . ... "Universality and structure of the N-end rule". J Biol Chem . 264 (28): 16700–12. PMID 2506181 . ^ a b c d Al-Dosari MS, Al-Muhsen S, Al-Jazaeri A, Mayerle J, Zenker M, Alkuraya FS (July 2008).
-
Testicular Germ Cell Tumor
Omim
In 1, the proband presented with a seminoma at the age of 51, his brother had had a testicular teratoma at the age of 28, and their cousin had an endodermal sinus tumor of the ovary diagnosed at 32 years. In the second family, the index case presented with an undifferentiated malignant teratoma at 28 years of age and his sister was diagnosed with bilateral mature teratomatous cysts at the age of 39. ... Kakinuma et al. (2001) found loss of heterozygosity at chromosome 1p in 21 (42%) of 49 Japanese TGCTs, including 12 (43%) of 28 seminomas and 8 (38%) of 21 nonseminomatous GCTs.
-
Gulf War Syndrome
Wikipedia
Persian Gulf War Illness Other names Persian Gulf War illnesses, and chronic multisymptom illness [1] [2] Summary of the Operation Desert Storm offensive ground campaign, February 24–28, 1991, by nationality Symptoms Vary somewhat among individuals and include fatigue, headaches, cognitive dysfunction, musculoskeletal pain, insomnia, [3] and respiratory, gastrointestinal, and dermatologic complaints Causes Toxic exposures during the 1990–91 Gulf War Differential diagnosis Chronic fatigue syndrome / myalgic encephalitis (CFS/ME); fibromyalgia ; multiple sclerosis (MS) Frequency 25% to 34% of the 697,000 U.S. troops of the 1990–91 Gulf War Gulf War syndrome or Gulf War illness is a chronic and multi-symptomatic disorder affecting returning military veterans of the 1990–1991 Persian Gulf War . [4] [5] [6] A wide range of acute and chronic symptoms have been linked to it, including fatigue , muscle pain , cognitive problems, insomnia, [3] rashes and diarrhea . [7] Approximately 250,000 [8] of the 697,000 U.S. veterans who served in the 1991 Gulf War are afflicted with enduring chronic multi-symptom illness, a condition with serious consequences. [9] From 1995 to 2005, the health of combat veterans worsened in comparison with nondeployed veterans, with the onset of more new chronic diseases, functional impairment, repeated clinic visits and hospitalizations, chronic fatigue syndrome -like illness, posttraumatic stress disorder , and greater persistence of adverse health incidents. [10] Exposure to pesticides and pills containing pyridostigmine bromide (used as a pretreatment to protect against nerve agent effects) has been found to be associated with the neurological effects seen in Gulf war syndrome. [11] [12] Other causes that have been investigated are sarin , cyclosarin , and emissions from oil well fire , but their relationship to the illness is not as clear. [11] [12] Studies have consistently indicated that Gulf War syndrome is not the result of combat or other stressors and that Gulf War veterans have lower rates of posttraumatic stress disorder (PTSD) than veterans of other wars. [9] [11] According to a 2013 report by the Iraq and Afghanistan Veterans of America , veterans of the U.S. wars in Iraq and Afghanistan may also suffer from Gulf War syndrome, [13] though later findings identified causes that would not have been present in those wars. [11] [12] Contents 1 Signs and symptoms 1.1 Comorbid illnesses 2 Causes 2.1 Earlier considered potential causes 2.1.1 Depleted uranium 2.1.2 Pyridostigmine bromide nerve gas antidote 2.1.3 Organophosphates 2.1.3.1 Organophosphate pesticides 2.1.3.2 Sarin nerve agent 2.2 Less likely causes 2.2.1 Oil well fires 2.2.2 Anthrax vaccine 2.2.3 Combat stress 3 Pathobiology 3.1 Chronic inflammation 4 Diagnosis 4.1 Classification 4.2 Kansas definition 4.3 CDC definition 5 Treatment 6 Prognosis 7 Prevalence 8 Research 9 Controversies 9.1 Jones controversy 10 Related legislation 11 See also 12 References 13 External links Signs and symptoms [ edit ] According to an April 2010 U.S. ... UK Australia Denmark Fatigue 23% 23% 10% 16% Headache 17% 18% 7% 13% Memory problems 32% 28% 12% 23% Muscle/joint pain 18% 17% 5% 2% (<2%) Diarrhea 16% 9% 13% Dyspepsia /indigestion 12% 5% 9% Neurological problems 16% 8% 12% Terminal tumors 33% 9% 11% * This table applies only to coalition forces involved in combat . ... PB had been used since 1955 for patients suffering from myasthenia gravis with doses up to 1,500 mg a day, far in excess of the 90 mg given to soldiers, and was considered safe by the FDA at either level for indefinite use and its use to pre-treat nerve agent exposure had recently been approved. [28] Given both the large body of epidemiological data on myasthenia gravis patients and follow-up studies done on veterans it was concluded that while it was unlikely that health effects reported today by Gulf War veterans are the result of exposure solely to PB, use of PB was causally associated with illness. [9] However, a later review by the Institute of Medicine concluded that the evidence was not strong enough to establish a causal relationship. [29] Organophosphates [ edit ] Organophosphate-induced delayed neuropathy (OPIDN, aka organophosphate-induced delayed polyneuropathy) may contribute to the unexplained illnesses of the Gulf War veterans. [30] [31] Organophosphate pesticides [ edit ] The use of organophosphate pesticides and insect repellents during the first Gulf War is credited with keeping rates of pest-borne diseases low. ... Department of Defense , "GWI is estimated to have affected 175,000 to 250,000 of the nearly 700,000 troops deployed to the 1990–1991 GW theater of operations. Twenty-seven of the 28 Coalition members participating in the GW conflict have reported GWI in their troops. ... Army Times . USA Today . Retrieved 28 January 2013 . ^ a b c d e "Pre 2014 Archived Meetings and Minutes, Research Advisory Committee on Gulf War Veterans' Illnesses" .
-
Arsenic Poisoning
Wikipedia
Histological observations confirm effects on cellular integrity, shape and locomotion. [28] DMA(III) is able to form reactive oxygen species (ROS) by reaction with molecular oxygen. ... Short-term arsenic exposure has effects on signal transduction inducing heat shock proteins with masses of 27, 60, 70, 72, 90, and 110 kDa as well as metallotionein, ubiquitin, mitogen-activated [MAP] kinases, extracellular regulated kinase [ERK], c-jun terminal kinases [JNK] and p38. [28] [103] Via JNK and p38 it activates c-fos, c-jun and egr-1 which are usually activated by growth factors and cytokines. [28] [104] [105] The effects are largely dependent on the dosing regime and may be as well inversed. ... "Dose-response relationship between prevalence of cerebrovascular disease and ingested inorganic arsenic". Stroke . 28 (9): 1717–23. doi : 10.1161/01.STR.28.9.1717 . ... "Cytogenetic effects in human exposure to arsenic". Mutation Research . 386 (3): 219–28. doi : 10.1016/S1383-5742(97)00009-4 .CCL20, AQP9, SKIL, SOD2, SRP68, SSBP1, TRAPPC10, TNF, TNFAIP6, TP53, UBE2E1, PIAS1, USO1, IER3, USP13, CCRL2, CD83, MINPP1, AKAP9, ZNF267, N4BP2L2, RUFY3, TNIK, TRA2A, SOX18, ZFAND6, ZNF331, PELI1, AS3MT, TAF1D, MIR145, SFPQ, CCL3L3, CCL4, ATXN7, CD44, CRP, ERCC1, ERCC3, ERCC4, GOLGA4, CXCL2, CXCL3, HSPA1B, ID2, IL1A, IL1B, IL1RN, INPP5A, KCNJ2, KRT10, MT1A, MTHFR, GADD45B, NDUFB8, PNP, NR4A2, PDE4B, PFKFB3, PTX3, RFX3, RGS1, GSR, ALAD
-
Community-Acquired Pneumonia
Wikipedia
Hospitalized patients have an average mortality rate of 12 percent, with the rate rising to 40 percent for patients with bloodstream infections or those who require intensive care. [28] Factors increasing mortality are identical to those indicating hospitalization. ... "Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP)" . MMWR Recomm Rep . 48 (RR–4): 1–28. PMID 10366138 . ^ Hayden FG, Atmar RL, Schilling M, et al. ... PMID 1462653 . ^ Ramirez, Julio A; Wiemken, Timothy L; Peyrani, Paula; Arnold, Forest W; Kelley, Robert; Mattingly, William A; Nakamatsu, Raul; Pena, Senen; Guinn, Brian E (2017-07-28). "Adults Hospitalized With Pneumonia in the United States: Incidence, Epidemiology, and Mortality" .CRP, COPD, TNF, IL6, ACE, IL10, HMGB1, MBL2, MMP9, ALB, TLR4, ADM, LNPEP, IRF5, SFTPB, SERPINB6, IL1RN, SLC9A6, HACD1, GRN, SLC2A10, SORBS1, BRD4, CTAA1, ANGPT2, COL4A5, CAP1, AVP, DELEC1, SDC4, NLRP3, SFTPD, SMPD1, TIMP1, TLR9, FGF21, HAVCR1, VDR, ARHGEF5, BHLHE40, BECN1, TIMELESS, SAFB, RGS6, RTN3, CCL18, A2M, RASA1, RARB, AGTR1, AMBP, BAG1, CAMP, CFTR, CHI3L1, DDX3X, FCGR2A, FCN2, HSPA1A, HSPA1B, HSPA2, TNC, IL1B, IL4, CXCL8, IL13, LTA, MCL1, MEFV, MRC1, MUC1, MYLK, SERPINE1, PTX3, MIF
-
Stevens–johnson Syndrome
Wikipedia
Drugs discontinued more than 1 month prior to onset of mucocutaneous physical findings are highly unlikely to cause SJS and TEN. [8] SJS and TEN most often begin between 4 and 28 days after culprit drug administration. [8] A published algorithm (ALDEN) to assess drug causality gives structured assistance in identifying the responsible medication. [13] [16] SJS may be caused by the medications rivaroxaban , [17] vancomycin , allopurinol , valproate , levofloxacin , diclofenac , etravirine , isotretinoin , fluconazole , [18] valdecoxib , sitagliptin , oseltamivir , penicillins , barbiturates , sulfonamides , phenytoin , azithromycin , oxcarbazepine , zonisamide , modafinil , [19] lamotrigine , nevirapine , [8] pyrimethamine , ibuprofen , [20] ethosuximide , carbamazepine , bupropion , telaprevir , [21] [22] and nystatin . [23] [24] Medications that have traditionally been known to lead to SJS, erythema multiforme, and toxic epidermal necrolysis include sulfonamide antibiotics, [8] penicillin antibiotics, cefixime (antibiotic), barbiturates (sedatives), lamotrigine , phenytoin (e.g., Dilantin ) ( anticonvulsants ) and trimethoprim. ... Similar to NSAIDs, paracetamol (acetaminophen) has also caused rare cases [27] [28] of SJS. People with systemic lupus erythematosus or HIV infections are more susceptible to drug-induced SJS. [10] Infections [ edit ] The second most common cause of SJS and TEN is infection, particularly in children. ... GARD . Archived from the original on 28 August 2016 . Retrieved 26 August 2016 . ^ a b c d "Orphanet: Toxic epidermal necrolysis" . ... S2CID 333724 . ^ Uzzaman A, Cho SH (2012). "Chapter 28: Classification of hypersensitivity reactions".HLA-B, HLA-A, HLA-C, IFNG, PTGER3, NOS2, IFNA2, MIF, NEDD4, NUCB1, ALB, EP300, PML, PSMC5, PTGIS, RB1, PARP1, RBX1, CAV1, DERL1, FBXO6, COPS5, CELF2, CTNNB1, VCP, ELMO1, CUL1, CUL4A, CSF3, NFKBIZ, POU5F1, TCF19, PSORS1C1, UMAD1, PSORS1C3, DDX39B, ATP6V1G2-DDX39B, GNLY, HSPG2, HLA-DRB1, TNF, TLR3, IKZF1, CYP2C9, CASP8, RBM45, TP53, CCL27, EGFR, MPRIP, CCL17, CYP2B6, CYP2C19, GZMB, RIPK3, HCP5, FAS, HLA-DQB1, ANXA1, CD33, ABCG2, SPAG9, WASF1, PROM1, CD37, CRP, SLC17A5, MIR18A, BAG6, MIR214, ZNF35, ALOX5, AKR1B1, UPP1, MICA, KEAP1, CASP9, FASLG, C5AR1, PART1, ARIH1, CABIN1, CD274, SETX, MARCHF1, PLXNA3, SCO2, TRAF3IP2, APC, PANK2, ABCB6, MLKL, BCL2L10, GSTM1, THBS1, TNFRSF1B, TLR2, COX2, CXCL9, FDXR, MDM2, LMNA, JAK2, IL15, IL13, IL10, IL4R, IL4, FPR1, IFNA13, FSHB, IFNA1, HMGB1, GFER, GJA1, CCR10, CFH, HDLBP, ETS2, ERCC1, NOS3, RET, TGFB2, TGFB1, SPTBN2, SOD1, SGSH, SFRP1, SDHD, SCT, SCN1A, CYP2C18, NPY, CYP2D6, CYP3A4, TIMM8A, DSP, EPHX1, ABCB1, PCM1, PAEP, ERBB2, PLG
-
Rickets
Wikipedia
Conversely, latitudes near the equator have selection for darker skin that can block the majority of UV radiation to protect from toxic levels of vitamin D, as well as skin cancer. [28] An anecdote often cited to support this hypothesis is that Arctic populations whose skin is relatively darker for their latitude, such as the Inuit, have a diet that is historically rich in vitamin D. ... PMID 15336592 . ^ Daily Telegraph , page 4, Wednesday 19 January 2011 ^ Rise in rickets linked to ethnic groups that shun the sun The Independent , published 2011-07-25, accessed 2011-07-251 ^ Doctors fear rickets resurgence BBC , published 2007-12-28, accessed 2011-07-25 ^ Loomis, W.G. ... PMID 12671133 . ^ National Health Service of England > Rickets Last reviewed: 28 January 2010 ^ "Children who drink non-cow's milk are twice as likely to have low vitamin D" . ^ "Rickets and osteomalacia" . nhs.uk .
-
Hypoxia (Medical)
Wikipedia
As the divers are decompressed , the breathing gas must be oxygenated to maintain a breathable atmosphere. [28] Inert gas asphyxiation may be deliberate with use of a suicide bag . ... NobelPrize.org . Retrieved 2019-10-28 . ^ "Hypoxia | GeneTex" . www.genetex.com . Retrieved 2019-10-28 . ^ a b Robinson, Grace; Strading, John; West, Sophie (2009).ACADVL, ESD, GAPDH, TFPI, SOD2, GNAS, ACTB, RPS14, MPO, NME1, NOS1, NOS3, NPPA, RPS2, PCK2, PDYN, PLAT, TKT, POMC, CA9, APOA1, ADK, SARDH, CTRB1, AHCY, ALDH1L1, CS, ALDOB, CPA2, EGR1, BDNF, ATP5F1B, ATF4, RET, ZFPM2, SCN8A, SLC35A1, PHOX2B, DNAH11, H3-3A, OAS1, MECP2, HBB, GDNF, GATA6, FBN1, ENG, EDN3, CSF2RB, CSF2RA, ASCL1, MAGEL2
-
Esophageal Cancer
Wikipedia
., and a low intake of fresh foods. [2] Other associated factors include nutritional deficiencies , low socioeconomic status , and poor oral hygiene . [16] Chewing betel nut (areca) is an important risk factor in Asia. [4] Physical trauma may increase the risk. [24] This may include the drinking of very hot drinks. [3] Adenocarcinoma [ edit ] Esophageal cancer (lower part) as a result of Barrettʼs esophagus Male predominance is particularly strong in this type of esophageal cancer, which occurs about 7 to 10 times more frequently in men. [25] This imbalance may be related to the characteristics and interactions of other known risk factors, including acid reflux and obesity . [25] The long-term erosive effects of acid reflux (an extremely common condition, also known as gastroesophageal reflux disease or GERD) have been strongly linked to this type of cancer. [26] Longstanding GERD can induce a change of cell type in the lower portion of the esophagus in response to erosion of its squamous lining . [26] This phenomenon, known as Barrett's esophagus , seems to appear about 20 years later in women than in men, possibly due to hormonal factors . [26] Having symptomatic GERD or bile reflux makes Barrett's esophagus more likely, which in turn raises the risk of further changes that can ultimately lead to adenocarcinoma. [16] The risk of developing adenocarcinoma in the presence of Barrett's esophagus is unclear, and may in the past have been overestimated. [2] Being obese or overweight both appear to be associated with increased risk. [27] The association with obesity seems to be the strongest of any type of obesity-related cancer , though the reasons for this remain unclear. [28] Abdominal obesity seems to be of particular relevance, given the closeness of its association with this type of cancer, as well as with both GERD and Barrett's esophagus. [28] This type of obesity is characteristic of men. [28] Physiologically, it stimulates GERD and also has other chronic inflammatory effects. [26] Helicobacter pylori infection (a common occurrence thought to have affected over half of the world's population) is not a risk factor for esophageal adenocarcinoma and actually appears to be protective. ... Archived (PDF) from the original on 29 April 2014 . Retrieved 28 April 2014 . ^ Vega, Kenneth J.; Jamal, M.CDKN2A, EGFR, PTGS2, WWOX, TP53, LZTS1, DCC, RNF6, TGFBR2, ADH1B, ALDH2, CASP8, MIR21, ERBB2, BCL2, CCND1, MIR145, ABCB1, XIAP, PRDX1, RUNX3, MET, LGALS3, PRDX2, SOX2, MMP14, KDR, XRCC3, BAX, SOD2, SERPINB3, MIR200C, TPM1, CDH13, MLH3, NR1I2, WIF1, CXCL2, ABL1, ACTB, ZNF667-AS1, MARCHF8, ALOX15, SFN, MIR98, PTGS1, GAPDH, GNG7, TNFRSF10A, CCNH, UCHL1, TPM4, GRIK2, CYP19A1, CYP26A1, PHB, MAP3K3, TGM2, MT1G, SST, CSF3, NOS2, CRYAB, SNAI2, NOS3, TUFM, ENO1, AKAP13, SMYD2, SLC30A7, MACIR, BCL2A1, AQP3, RPRM, TRMT11, GHRL, BBC3, ADH7, MYC, IL5, CXCL1, MXI1, HMOX1, CTNNA1, PLCE1, STK11, MUC1, RHBDF2, SLC52A3, HLA-A, RUNX1, SLC39A6, ADH4, CHEK2, PCAT1, ERCC1, FKBP1A, MGMT, FLACC1, PEBP4, TMC1, MTHFR, LINC01504, MTAP, CCDC63, DGKH, GSTM1, SDCBP2-AS1, TSPAN11, STEAP1B, MCC, HECTD4, KIT, TEX41, IL6, LINC01405, OCA2, PLCE1-AS1, SLC25A24P1, GSTT1, GSTP1, JAZF1, PIK3CD, PDE4D, TBL2, TFDP1, HEATR3, SEMA5B, TNF, TRPC4, CD274, XBP1, ZFAND5, STAT3, CUBN, SMUG1, IL18R1, PES1, SMG6, KANK1, IKZF3, ABCC8, SREBF2, PDGFRA, PTPN2, SGF29, PIK3CA, PIK3CB, PIK3CG, MAPK1, PTEN, AGBL4, SPINK7, ACAD10, ST6GAL2, SDHA, SDHB, SDHC, ST6GAL1, FAM167A-AS1, FAM167A, VEGFA, H3P10, CYP2E1, AKT1, CDH17, AMPH, CLN3, CRP, CSNK1A1, CTNNB1, AHR, CYP1A1, CALR, CALCR, MDM2, CEACAM5, SLC6A8, HIF1A, TGFB1, NQO1, MIR203A, COX2, EGF, EPHX1, CDKN1A, APC, XRCC1, IGF1, VEGFC, AKR1A1, EPHB2, FHIT, PRM3, MIR34A, RAD51, RARB, IL10, UCA1, TERT, MIR375, IGF2, MMP2, TP73, MTCO2P12, GLI1, CXCR4, PPARG, CCK, EPCAM, MMP7, BMI1, IGFBP3, PCNA, RASSF1, HPSE, OGG1, DNMT1, TMPRSS11A, HPGDS, ECRG4, YAP1, SLC12A9, CDH1, TWIST1, BSG, NAT2, FGF2, EZH2, EGR1, DPYD, IGF1R, ARHGAP24, E2F1, CDK2, MMP9, AURKA, MALAT1, PDCD4, COMMD3-BMI1, BRCA2, FAS, IL1B, MIR93, MIR143, IL18, CDKN2B, TLR4, CKAP4, S100A4, MAPK3, KRAS, FOXA1, PDPK1, ERCC2, ALB, TP63, TNFSF10, ALDH1A1, IFNG, SOX4, PROX1, BHLHE40, CTAG1A, IL1RN, CST3, SOX17, CXCL8, AVP, TYMS, SLCO6A1, CTAG1B, IDO1, VDR, GSTK1, CASP3, BAGE4, PRKCI, CTTN, MIR141, CASC9, PDK1, GNAS, ATAD1, AR, LATS2, MIR139, ADH1C, LOXL2, CDK4, ZNRD1, MIR10B, DELEC1, PLCL1, ABCG2, HMGB1, CYP2A6, LINC01194, MIR214, TERC, CDKN1B, MIR155, ORAI1, PLK1, CXCL12, SERPINB4, CEACAM3, F2RL1, BIRC5, FOXQ1, NKD2, SKIL, CDKN3, SKP2, FASLG, CCNB1, IL27, IL23R, PRKAA2, LGMN, MIRLET7C, MIR100, MIR107, PRKCA, PRKAB1, PRKAA1, PSMB8, POU5F1, POLR2E, POLB, PLAU, MIR144, PKM, PSG2, PTK2, PAQR3, CEACAM7, ANXA2, RPS6KB1, ANXA1, ANGPT1, RPE65, DACT2, RNF2, PTK7, REG1A, RBBP6, RB1, FAP, RARA, SLC39A5, HAVCR2, CDC34, SOX9, ZEB2, CTAG2, CRNN, GEMIN4, CLOCK, KLF4, PTTG1, MTA1, BCL2L1, IL17D, HSPB3, BAG1, SPHK1, RUNX2, KAT2B, PROM1, CCN4, UGT1A1, IST1, BRCA1, SPP1, NXT1, NLRP1, RIPK3, DKK1, RAB40B, SUB1, TBC1D9, ENG, SIRT1, MRPL28, SNHG1, NDRG1, TUBB3, PSMD14, BUB1B, UBE2S, CHAF1A, FRAT1, ERBB4, TNFRSF10B, FEV, LY6K, ANKRD36B, EIF4EBP1, GAS5, TTK, TRAF6, BHLHE41, TP53BP1, CDC25B, TNFAIP3, ZC3H12A, TGM3, PDCD1LG2, CLPTM1L, EIF4E, CDK6, AFAP1-AS1, SRC, UNG, UVRAG, CD38, FERMT1, AXL, ZNF654, FBXW7, AXIN2, MEG3, ADAM12, TUSC3, TENM3, WNT1, MYDGF, DIABLO, XRCC4, EYA4, XK, CD34, TP73-AS1, PIN1, SATB1, MIR146A, MIR29C, IL17A, CXCL10, IRF2, MIR148A, JUN, JUNB, JUND, POU5F1P4, POU5F1P3, KRT16, MIR483, MIR193B, SMAD4, MIR451A, MAGEA3, MAL, CTSB, MDK, MKI67, MLH1, MIR377, ABCC1, MSH2, FGF13, MIR301A, MIR576, IL12B, CXCR2, HGF, H3P9, GJB2, GML, GRB7, CCAT2, FALEC, GSN, GSTM2, GSTM3, H2AX, HDAC1, MTOR, IL2, UBE2K, ADAMTS9-AS2, CD24, HRAS, HSPB1, HSPB2, HSP90AA1, HOTAIR, FOSB, FOS, IL1A, MAPK14, SNORD116@, CASP10, MIR199A1, MIR199A2, MIR183, PEBP1, ODC1, NQO2, CRYZ, FGF7, NRDC, MIR27A, FBXL19, UGT1A8, MIR200A, GAEC1, DLL4, AKT2, MIR20A, MIR873, MARCHF5, OTUD4, MIR122, MIR543, CRCT1, KRT20, TLR9, PARP1, SIRT6, NT5C3A, SLC25A37, DACT1, LINC00184, CHST15, ZMYND10, UBR5, IL23A, SYCE1L, MIR1294, ELK1, MIR126, SPA17, ADAR, XAF1, NANS, TDRG1, FBLIM1, NBAT1, CT47A4, SAGE1, SOX6, MIR206, OTUB1, DSG3, CT47A5, MFN1, CT47A6, CHFR, ACSS2, CT47A8, ERBIN, CT47A9, STK31, CT47A10, MIR210, BDH2, ZNF331, TMEM176A, HULC, LAPTM4B, ACP6, TRIM44, RNF111, SYTL2, MAGED4, TUG1, CT47A1, MIR204, SLC52A1, ANO1, SBNO1, NAT10, QRSL1, MIR205, CT47A2, CHDH, CT47A3, IGF2-AS, LINC01234, PANTR1, BNIP3, CSAG2, SEMA3B-AS1, LINC01617, KCNH4, SLC44A1, ANGPTL2, SLC25A20, SEC14L2, PADI4, CA2, MIR186, PLA2G15, TMEFF2, C4BPA, LINC-ROR, MIR187, PRDX5, MPRIP, CERNA3, PEG10, SBNO2, OIP5, RRAS2, CASP1, PUF60, H3P17, ZHX2, MIR150, MMRN1, MIR16-2, RAB3GAP1, MIR152, TPX2, TPM1-AS, DNMT3B, KDM1A, SMG1, RNF19A, MTHFD1L, POLDIP2, IL22, MIR197, PSAT1, CT47A12, MIR129-2, MIR198, STOML2, MIR127, CYHR1, MIR140, CCAT1, DDX11-AS1, F11R, FOXP3, NDUFA13, LEF1, ATP6V0C, HOOK2, ACTA2, ZBTB20, EMP3, RGS22, PHGDH, NKX2-8, ABO, DNM3OS, MIR550A3, PABPC1, LAMP3, DROSHA, MIR192, IL37, RPL17-C18orf32, EMP1, MIR196A1, MIR196A2, ACACA, C1GALT1, MIR296, ELANE, RHOXF2, MIR423, TSPAN18, EMILIN3, ABCB5, DHCR24, WDR20, MIR382, FFAR4, MTDH, SCGB3A1, NAF1, MIR361, PRRT2, MIR374A, MIR373, FBXO32, APEX1, TSLP, DUXAP8, BAGE3, PWAR4, MIR494, ARG1, MIR202, MIR146B, ACCS, CCDC54, MIR491, RIOX2, DUXAP10, SOX2-OT, MIR433, VSIG1, AQP4, MIR20B, MIR363, SNHG7, MIR372, KLF17, OSR1, JMJD1C, ZFP82, ATN1, MIR96, ADAMTS16, MIR34B, DSC2, MIR31, ALOX15B, CBLL2, FOXK1, MIR30E, MIR30B, MIR302A, CT47A11, ALOX5, MIR24-1, MIR17HG, LRATD2, EPHA2, LINC00261, KCNH8, MIR340, CKS1BP7, CTCFL, TMED10P1, RPL34-AS1, MIR339, OR2AG1, AGER, DACT3, ANXA7, MIR338, PDIK1L, MIR335, MIR326, MIR302B, LINC-PINT, MIR498, CRISP1, MIR499A, USP28, KIAA1522, MIR574, SUGP1, MIR556, IL21, RERE, ZNF667, MIR219A1, GOLPH3, MIR542, MIR539, GORASP1, CDCP1, MARCKSL1, RSRC2, VLDLR-AS1, MIR212, KLHL1, MIB1, SLC44A2, ADAMTS9, IS1, ACKR3, MIR655, AKR1B10, MIRLET7G, CYSLTR2, MIR625, POTEG, MIR601, DANCR, S100A14, NDRG2, TENM2, INTS2, MIR593, WNK1, TRPM8, FSD1, MIR506, MIR487A, ZFHX3, MIR486-1, MAGED4B, NECTIN4, TSC22D4, MIR224, MIR502, ALPK1, ARSA, ARNTL, CRISPLD2, USP26, FSD1L, PHF6, SARNP, WNT10A, RNF34, ALCAM, VTCN1, MUL1, CT47A7, FAT4, DPP4, MIR221, ATM, NEIL1, LIN28A, CSAG3, FBXO31, RASSF10, NANOG, WLS, GRHL2, NAA25, C17orf97, SSX2B, XRCC6, CAV1, PDHA1, CDK14, PGD, PGF, COL11A2, COL6A3, CNTN1, CNR1, PIM1, ABCC2, LTB4R, PLA2G2A, PLG, PRRX1, PON3, POR, POU2F1, POU5F1B, PPL, PRKCB, CCR5, MAPK8, MAP2K1, MAP2K3, PECAM1, PDGFRB, MAP2K7, CLDN4, TRIM37, CSF2, MYD88, GADD45B, NF2, NGF, NOTCH1, NPAS2, CRMP1, FGF4, SLC22A18, CRK, PAH, SERPINE1, PRKN, PAWR, PAX2, PBX1, FGF3, PCYT1A, PDCD1, PDE4A, CREB1, MAP2K6, EIF2AK2, FGF12, RMRP, RPA1, RPL15, RPL17, RPL34, RPN2, RRAD, RRAS, S100A9, SAFB, SALL2, SAI1, TSPAN31, SAT1, CCL1, CCL2, CCL11, CCL15, CCL18, CCL20, CXCL11, CFTR, CEBPB, F3, ABCE1, RIT1, HTRA1, RELB, KLK10, PSMB9, PSMD4, PTCH1, FEN1, CKS1B, CHRNA3, CHGA, PTPN6, PVT1, RAC1, RAC3, RAD9A, RAD23B, FASN, RAF1, RAP1A, RARRES2, RBL2, RBM3, RBM4, RBP2, RELA, MUC4, MTR, DDX5, FOXA2, HOXA1, HOXA9, HOXA13, HOXD13, HP, HPGD, ERAS, HSPG2, ICAM1, ICAM2, ICAM3, ID1, IFNB1, IFNGR1, IGFBP1, FLT1, CCN1, IL3, CYP1B1, FLNB, IL7R, IL9, FOXO1, FOXA3, HLA-G, IL12A, HLA-DRB1, GAS6, GJA1, G6PD, GLI2, GNA12, DCK, DAPK1, GPR39, GRN, DAP, NR3C1, DACH1, DAB2, GRP, GSK3B, GSR, CYP24A1, FZD2, CYP2C9, HDGF, CYP2C19, HLA-DOA, HLA-DQA1, IL11, IL12RB1, MTTP, MCM7, FOXC1, CTNNA2, MFGE8, CCN2, MAP3K11, MMP1, ACE, FGFR4, MMP10, MMP13, CTBP2, MPO, MSH3, MSI1, MST1, MT1E, MT2A, MT3, CSTA, CYTB, MTHFD1, ND4, ND5, FOXC2, SMAD7, IL13, SMAD5, IL15, IL15RA, PDX1, ITGA6, IVL, JAK1, JAK2, CYLD, CXADR, KRT1, KRT7, KRT10, KRT19, STMN1, CTSL, LGALS9, LLGL2, LLGL1, LMX1B, LOX, TM4SF1, FOXM1, MAD2L1, SEL1L, SHH, SHMT2, F2RL3, GPRC5A, PIAS2, CLDN1, CCNE2, DCLK1, AURKB, NOLC1, RAB11B, IL32, NOG, MSC, UBE2L6, DCN, GRAP2, MAP4K4, MAGED1, PTGES, CHD1L, AKAP12, ISG15, MDC1, PCLAF, RASSF2, MAP3K14, LIMD1, BMS1, BCL10, LTBP4, RECK, CUL4A, CUL2, PPM1D, IFITM1, STC2, USO1, AKR1C3, BECN1, URI1, RIPK1, TNFRSF14, TNFRSF6B, TNFRSF11A, SCEL, GADD45A, BANF1, NRP1, CCN5, HDAC3, CDK5R1, PER2, EIF4A3, MAML1, CDX2, NXF1, ARL6IP5, AHSA1, TXNIP, PDPN, POSTN, CELF1, PTGES3, HSPH1, EPHA3, JTB, GADD45G, FASTK, BTG3, MLLT11, TMED10, FERMT2, KCNQ1OT1, SPINK5, IL24, CNMD, CD160, FSTL1, CAV2, CIB1, DDIT3, SPATA2, RAPGEF3, FARP2, RBX1, HDAC6, PDCD6, PARP2, DNAJB6, ABCB6, DNM1L, TSPAN1, ARL4C, TRAP1, ARFRP1, G3BP1, GDF11, TRIB1, MSLN, AKAP8, BCAS2, APC2, PAK4, TNIP1, ERBB3, RACK1, FZD7, BAP1, AXIN1, TCF4, ZEB1, TCF21, TRBV20OR9-2, TDO2, TERF1, EVPL, CDH3, TGFA, ETV1, TGFBI, TGFBR1, ETS1, CDC42, THRB, THY1, TIMP1, TIMP2, TLE2, TSPAN7, TSPAN8, TM7SF2, TMPO, TMSB4X, TCF7L2, TAP1, TAM, TAGLN, SIX1, SIX3, SLC1A5, SLC3A2, SLC11A1, SLC22A1, SMARCA2, SMARCA4, SIGLEC1, SNAI1, SNCA, SNCG, FSCN1, SOAT1, CDKN1C, SPRR3, SSR1, F2R, SSX2, STAT1, STC1, SULT1A1, SYT1, TOP1, ERCC5, CDC25A, TPM2, WRN, XIST, XPC, CD80, XRCC5, YES1, YES1P1, PCGF2, ZBTB17, ZNF208, CD3E, PRDM2, IL1R2, MANF, USP7, ST8SIA4, AIMP2, TFPI2, CCNG2, HMGA2, SLC7A5, MIA, NRIP1, WNT2B, WNT10B, VRK1, CD59, CDC20, TPT1, TRH, CD63, TRPC6, TSC1, ABCA4, ERCC4, UBE2D3, UBE2E2, UCP1, VIM, SLC35A2, UGT1A, UGT2B4, UMOD, UMPS, USP4, NR1H2, ERCC3, VHL, EZR, MAPKAPK2
-
Guillain–barré Syndrome
Wikipedia
In the first two weeks, these investigations may not show any abnormality. [4] [20] Neurophysiology studies are not required for the diagnosis. [8] Formal criteria exist for each of the main subtypes of Guillain–Barré syndrome (AIDP and AMAN/AMSAN, see below), but these may misclassify some cases (particularly where there is reversible conduction failure) and therefore changes to these criteria have been proposed. [28] Sometimes, repeated testing may be helpful. [28] Clinical subtypes [ edit ] A number of subtypes of Guillain–Barré syndrome are recognized. [4] [28] Despite this, many people have overlapping symptoms that can make the classification difficult in individual cases. [5] [29] All types have partial forms.PMP22, CD86, AIRE, TNF, IL17A, CSF2, LAMC2, CD1E, ALB, CD1A, HLA-DQB1, IL1B, CD1C, TLR4, IL10, CD1B, ICAM1, IL23A, ISG20, FCGR2A, CRP, IL4, IL2RA, IL17D, HLA-DRB1, IL6, MAPK1, NFASC, GFAP, ITGAM, COX2, MYDGF, PTGS2, FCGR3A, APOE, IL27, FAS, MTCO2P12, CNTNAP1, YY1, YWHAZ, CXCR4, AIMP2, RETN, SELENBP1, TNFRSF1B, MIR155, GRAP2, GLDN, NOD1, VIM, MIR642B, SIGLEC14, AHSA1, OCLN, TLR2, TH, TGFB1, STAT3, SPP1, SMPD2, CXCL6, CCL2, PTPN11, MIF-AS1, IVNS1ABP, CSGALNACT1, POLDIP2, SMPD3, NS2, IL21, ISYNA1, FOXP3, IL22, ICOS, DLL1, ERVK-6, SIGLEC9, ST6GALNAC4, PTGS1, RNF19A, TUBGCP2, NOD2, SUMF2, FTSJ1, IL33, CADM1, FCRL3, FTSJ3, RBM45, HT, CABIN1, PLB1, YWHAQ, SIGLEC7, NOS2, PSMB6, F2R, HMGB1, HLA-DQB2, HLA-DQA1, HLA-DOA, CXCL2, CXCL1, NR3C1, GOLGA4, GNAO1, GLO1, GJB1, GALE, FOLH1, FCGR3B, ESR1, HPRT1, EPHB2, ENO2, CST3, MAPK14, CRK, COX8A, CNTN1, CDC42, CD59, CD80, CD14, CACNA1A, SERPING1, FASLG, HP, HSPA4, MAP2K7, MMP2, MAPK8, PRKD1, POMC, PLA2G1B, PKD1, PGF, PDCD1, NPPB, NPY, NOS3, NEFL, COX1, MRC1, MMP9, NR3C2, HSPD1, MIF, MBL2, LEP, ITGB2, ITGAL, IRF6, INSRR, CXCL10, IL18, IL12B, IL12A, IL2, IGHG3, IFNG, ERVK-32
-
Colorectal Cancer
Wikipedia
The classic warning signs include: worsening constipation , blood in the stool, decrease in stool caliber (thickness), loss of appetite, loss of weight, and nausea or vomiting in someone over 50 years old. [13] Around 50% of individuals with colorectal cancer do not report any symptoms. [14] Rectal bleeding or anemia are high-risk symptoms in people over the age of 50. [15] Weight loss and changes in a person's bowel habit are typically only concerning if they are associated with rectal bleeding. [15] [16] Cause [ edit ] 75–95% of colorectal cancer cases occur in people with little or no genetic risk. [17] [18] Risk factors include older age, male sex, [18] high intake of fat, sugar , alcohol , red meat , processed meats , obesity , smoking , and a lack of physical exercise . [17] [19] Approximately 10% of cases are linked to insufficient activity. [20] The risk from alcohol appears to increase at greater than one drink per day. [21] Drinking 5 glasses of water a day is linked to a decrease in the risk of colorectal cancer and adenomatous polyps. [22] Streptococcus gallolyticus is associated with colorectal cancer. [23] Some strains of Streptococcus bovis/Streptococcus equinus complex are consumed by millions of people daily and thus may be safe. [24] 25 to 80% of people with Streptococcus bovis/gallolyticus bacteremia have concomitant colorectal tumors. [25] Seroprevalence of Streptococcus bovis/gallolyticus is considered as a candidate practical marker for the early prediction of an underlying bowel lesion at high risk population. [25] It has been suggested that the presence of antibodies to Streptococcus bovis/gallolyticus antigens or the antigens themselves in the bloodstream may act as markers for the carcinogenesis in the colon. [25] Inflammatory bowel disease [ edit ] People with inflammatory bowel disease ( ulcerative colitis and Crohn's disease ) are at increased risk of colon cancer. [26] [27] The risk increases the longer a person has the disease, and the worse the severity of inflammation. [28] In these high risk groups, both prevention with aspirin and regular colonoscopies are recommended. [29] Endoscopic surveillance in this high-risk population may reduce the development of colorectal cancer through early diagnosis and may also reduce the chances of dying from colon cancer. [29] People with inflammatory bowel disease account for less than 2% of colon cancer cases yearly. [28] In those with Crohn's disease, 2% get colorectal cancer after 10 years, 8% after 20 years, and 18% after 30 years. [28] In people who have ulcerative colitis, approximately 16% develop either a cancer precursor or cancer of the colon over 30 years. [28] Genetics [ edit ] Those with a family history in two or more first-degree relatives (such as a parent or sibling) have a two to threefold greater risk of disease and this group accounts for about 20% of all cases. ... Patients with positive regional lymph nodes (any T, N1-3, M0) have an average five-year survival rate of approximately 40%, while those with distant metastases (any T, any N, M1) have an average five-year survival rate of approximately 5% and an average survival time of 13 months. [144] [145] Epidemiology [ edit ] Colon and rectum cancer deaths per million persons in 2012 3–17 18–21 22–27 28–36 37–54 55–77 78–162 163–244 245–329 330–533 Globally more than 1 million people get colorectal cancer every year [18] resulting in about 715,000 deaths as of 2010 up from 490,000 in 1990. [146] As of 2012 [update] , it is the second most common cause of cancer in women (9.2% of diagnoses) and the third most common in men (10.0%) [12] : 16 with it being the fourth most common cause of cancer death after lung , stomach , and liver cancer . [147] It is more common in developed than developing countries. [148] Globally incidences vary 10-fold with highest rates in Australia, New Zealand, Europe and the US and lowest rates in Africa and South-Central Asia. [149] United States [ edit ] Colorectal cancer is the second highest cause of cancer occurrence and death for men and women in the United States combined. ... United Kingdom [ edit ] In the UK about 41,000 people a year get colon cancer making it the fourth most common type. [152] Australia [ edit ] One in 19 men and one in 28 women in Australia will develop colorectal cancer before the age of 75; one in 10 men and one in 15 women will develop it by 85 years of age. [153] History [ edit ] See also: Timeline of colorectal cancer Rectal cancer has been diagnosed in an Ancient Egyptian mummy who had lived in the Dakhleh Oasis during the Ptolemaic period . [154] Society and culture [ edit ] Main article: List of people diagnosed with colorectal cancer In the United States, March is colorectal cancer awareness month . [97] Research [ edit ] Preliminary in-vitro evidence suggests lactic acid bacteria (e.g., lactobacilli , streptococci or lactococci ) may be protective against the development and progression of colorectal cancer through several mechanisms such as antioxidant activity, immunomodulation , promoting programmed cell death , antiproliferative effects , and epigenetic modification of cancer cells. [155] Mouse models of colorectal and intestinal cancer have been developed and are used in research. [156] [157] [158] The Cancer Genome Atlas [47] The Colorectal Cancer Atlas integrating genomic and proteomic data pertaining to colorectal cancer tissues and cell lines have been developed. [159] References [ edit ] ^ a b c d e "General Information About Colon Cancer" . ... Archived from the original on September 28, 2015. ^ Juul JS, Hornung N, Andersen B, Laurberg S, Olesen F, Vedsted P (August 2018).MLH1, NRAS, APC, MSH2, DCC, MMP2, SMAD4, PIK3CA, SMAD3, CHEK2, BRAF, CTNNB1, TCF7L2, BUB1, TP53, FGFR3, KLF5, EP300, KDR, MSH6, IGFBP3, AKT1, ABCB1, KRAS, RET, POLD1, POLE, STK11, NAMPT, SLCO1B3, EYA4, BUB1B, SYNE1, EVL, ABCA1, LGR6, ABCB5, DACT1, PRKD1, TCERG1L, IL6ST, PRKCE, PMS2, COX1, COX2, SH3TC1, GUCY1A2, RNF182, PPARG, MLH3, AXIN2, TGFB1, BMP4, GREM1, MCC, CD109, CDH1, PTPRD, SMAD7, HAPLN1, MUTYH, ARID1A, GNAS, DLC1, CCND2, RHPN2, FEN1, LRP1, LPAR1, SLCO2A1, VTI1A, HHIP, TET2, POLD3, TMEM238L, LAMC1, PDGFRL, CUBN, ZMIZ1, NXN, PROM1, XRCC1, EGFR, FADS1, ERCC2, SMAD9, SFRP2, FLCN, PTEN, SFRP1, DCLK1, FADS2, CCND1, ABCG2, PTGS2, SMAD2, SPARC, BCL2, CXCL8, CALR, TYMS, IGF2, TFPI2, CYP1A2, TNF, TLR4, BOC, TLR2, CASP8, MTDH, PARP1, TGFBR2, TGFBR1, CYP1B1, LGR5, FN1, SATB2, TNFSF10, ESR2, AURKA, SRC, WIF1, MYC, DPYD, YAP1, MMP1, CHD1, FBXW7, LEF1, ALDH2, RNF43, GSTM1, ABCC1, NQO1, ODC1, NME1, CYTB, GUCY2C, ND4L, KRT20, MTHFR, TYMP, MTRR, TGM2, ADH1B, NTHL1, BIRC5, TIAM1, ATG5, WNK1, EPHA1, ZEB2, TNFSF13, BMP2, CSF2, SOD2, RRM2, MIR26B, PRKCB, EXO1, MIR106B, AKAP12, FPGS, IDH1, KL, ING4, CSE1L, TLR9, GGH, AKR1B10, CDO1, MPO, MIR93, ACKR3, DHFR, SLC19A1, MMP11, WRN, TK1, GALNT12, KLF2, ABCC3, KMT2C, STRAP, NR4A2, PRKCZ, CHD5, AMACR, ABCC2, PPP2R1B, AKAP9, GATA5, ATAD1, MIR130B, GAPDH, FAT1, RPS6KB1, FOLH1, EPHA3, SESN2, SELENOP, ZNF217, MIR1271, TFRC, BMPR1A, MKI67, TP63, IRS4, CYP2A6, EIF3H, DPEP1, CAPN10, PDGFD, POSTN, TCN2, XAF1, TCF3, LIFR, CFTR, TPX2, PYCARD, PTPRU, LAMA1, ABCB6, CPE, EFEMP1, ARNT, RTN4, RASSF2, RASAL2, BRINP1, APC2, UMPS, SFRP4, EPHB6, SRSF6, TNS4, SELENBP1, PRIMA1, PER1, TENM1, ACSL4, IFNA1, NUSAP1, TRIM28, CTNND2, MTHFD1L, NTNG1, ABCC5, ASAP3, ABCC4, SLC12A5, SLC29A1, EPHA7, SOX17, CENPH, MCRS1, GPNMB, CD9, ZKSCAN3, ACSL5, FZD4, ERCC6, CUX1, CAD, GPX4, CACUL1, SCG5, GATA4, ADAMTS15, PTPRS, PRPS1, SERPINB5, BMP6, GSTM3, GTF2B, P2RX7, NME2, ACACA, ABCA4, TBX1, VWA2, RARB, LRP2, RASGRF2, APOB, RHEB, DDIT3, ACIN1, TXNRD1, GRIN2A, CHL1, FBLN2, HRH1, SFRP5, ATP7B, UNG, ZNF292, NEURL1, SLC11A2, HK3, CD226, FOXH1, GNB4, DHRS2, COL3A1, ADAMTSL3, PTPRT, NFATC1, GLI3, SDF4, CD248, NF1, FBN2, UQCRC2, NR3C2, CD93, CD46, ADORA1, EIF4G1, OBSCN, GHRHR, ADAMTS18, QKI, NCR1, GSE1, SKIV2L, SF3A1, DPYSL2, CDC14A, CTBP1, LIG3, PPP1R14A, F5, FOXL2, KDM6A, RUNX1T1, DMKN, TRPM7, OTOP2, CABLES1, TNFRSF9, ANP32A, BRF1, IFNA2, LY96, PCSK2, ST8SIA4, ABCB4, PRSS1, VPS13A, TTLL3, PKHD1, SETBP1, HDC, NRCAM, ADAM19, ABCB10, ODAM, LRRC3B, LDB1, UHRF2, CSMD3, OVGP1, GNB5, UNC13B, DPAGT1, OSBP, ADAM29, DNASE1L3, DLG3, ARFGEF1, MKRN3, DMD, ZNF155, DNAH8, NODAL, AKAP3, ALDH1L1, DSTN, FANCG, NID1, NELL1, CAMSAP2, LIG1, AGTPBP1, LAMB3, KCND3, KCNH4, ABCA6, ABCA5, KCNA10, CBX5, PIK3R5, HTR5A, ISLR, IPP, ZNF43, FCN1, RAB38, LMO7, LPIN1, NCDN, CNKSR2, NDUFA1, MYO1B, TP53TG1, TFEC, DEFA4, PPM1E, HOXC9, HOXD1, MCF2L2, H1-4, DIP2C, MAP2, HOXD9, SNRNP200, MAP1B, MAF, H1-5, CALCOCO2, ZNHIT1, WNT8B, SNRPB2, SLCO1A2, GABPB1, PAX8, MCM3AP, GABRA6, SMTN, SLC1A7, KALRN, ARHGEF1, GALNS, SLC33A1, ZMYM4, SCNN1G, KCNB2, SCNN1B, RPL6, ABCE1, RCN1, SORL1, SPTBN2, CDKL5, H2BC14, TUBB2A, STAM, FHL3, MLF2, ICAM5, RBM10, GFI1B, FLNC, OR1E2, SULT2B1, HAT1, TCOF1, DCHS1, ABCB11, ETV1, NPAP1, ETFDH, ABCC8, RBP3, RALGDS, DNALI1, BCAS2, PPOX, EDA, GRID1, GRID2, GRIK3, PMM2, GRM1, EFS, PKNOX1, PRG2, TFG, ABCA10, ABCA9, ABCA8, ZNF232, CORO2B, PGM1, PEG3, FCN2, PQBP1, AKAP6, STARD8, MYOT, RAB5C, ABCG1, PPM1F, ZNF432, PZP, ACAP1, CLSTN3, ABCD4, NPBWR2, ABCD3, JAKMIP2, PTPRN2, PSMC5, PSMA2, PAN2, P2RY14, PROS1, NDST3, KDM2A, EML2, ZNF442, CEBPA-DT, SLC66A2, PREX2, CBFA2T3, BPIFB2, TRAF3IP3, ZSCAN16, PBX4, SLC44A4, LAS1L, CAPN6, FBXO30, TOMM40L, TMEM164, HPS3, LZTS2, CCDC62, CACNB2, CACNB1, PIGO, LMNB2, IGSF21, CCNB3, FAM110B, ZNF804A, MYLK3, DSEL, OLFM2, ZFP36L2, CSRNP3, MMRN2, DSCAML1, ZNF385D, NLGN4X, ZNF624, ZNF471, RELCH, CDH7, CDH5, VPS18, HIVEP3, TSKS, EXOC4, DUSP21, DMRTA1, CLSTN2, LRRC4, NCAPG, MS4A5, CHST8, GMCL2, CDH22, ARAP3, PLPPR2, BCL11B, SLC26A10, BOLL, CYREN, GDAP1L1, ZSCAN5A, RIC3, PARP8, SFXN1, ERI2, OMA1, C8orf34, SPNS3, PDILT, AMER2, GJD4, FERD3L, ITPRID1, TTLL10, STK32C, ABCC6, ACTL9, ATP11C, KRT73, CD300E, KRTAP10-8, RASL11A, CCDC9B, CFAP77, C12orf76, AMTN, ANK2, AMPD1, ABCD2, ADORA3, ZBTB8B, ADCY8, ADARB2, MIR1273C, LINC00673, ABCA3, AMZ2P1, SPAG17, VSTM4, ZNF560, CSNK1A1L, SPACA7, B4GALNT2, CYB5D2, NLRP8, KDF1, CPO, EVC2, ZNF572, FRMPD2, SYT9, PHETA1, ZNF480, ZNF569, HTR3C, ABCC13, PUS10, C2CD6, ATP6V1E1, WDR49, CNTN4, JAKMIP1, ABCA13, IQUB, ERICH1, ZNF540, LONRF2, UGT3A2, LRRC47, SLC22A9, PAIP2, PANK4, ZNHIT6, CTSH, PHIP, EDRF1, ARHGEF10L, COL4A6, PCDH11X, VRTN, JPH3, TMEM140, SLC22A15, DNAH3, ZDHHC7, MFN1, INTS13, CCAR1, CINP, CCDC93, SCN3B, GPR87, AMDHD2, SHANK1, NT5C, DONSON, PDZRN4, ERGIC3, CNTN1, TSPEAR, ELOA2, ANAPC4, ZZZ3, SNX8, PEX5L, SETD4, TNNI3K, ETAA1, SALL3, TBX22, CPAMD8, FBXL2, WNT16, KCNQ5, ZDHHC2, SIGLEC7, FSTL5, ABCA12, PAK6, MFF, COL7A1, WAC, RPAP1, CABYR, CNPPD1, SDCBP2, HR, SACS, CORO1B, RNPEPL1, AGER, BAX, CD59, NFE2L2, BRCA2, CCAT2, CDH3, FGFR2, BCL10, SLC22A3, PTPRJ, STS, PLA2G2A, LRIG1, CASC8, COLCA2, HLA-B, NOS1, LAMA5, PCAT1, KRT8, MPRIP, SH2B3, COLCA1, CYP17A1, WWOX, ATF1, EDN1, BMP5, GATA3, UTP23, MZF1, NOTCH4, TMBIM1, CDKN2B-AS1, FMN1, FKBP5, RAD54B, RTEL1, ERAP1, RALY, SHROOM2, C11orf53, TNS3, ELOVL5, TNS1, ETV6, BRIP1, EPHA5, CHRDL2, TFEB, HLA-F, SBF2, HNF1B, HSPA12A, SYMPK, MIR193A, MIR195, MIR20A, MIR17, OGG1, NOTCH1, MIR200C, MIR200B, MIR203A, MIR183, MIR18A, MIR182, GPR143, NTRK1, NDRG2, MIR155, PMP22, PIK3CD, PIK3CG, PKM, PRPF31, PLAU, PLK1, ZBTB20, SIRT1, MALAT1, MIR150, GSTK1, TICAM2, SLC6A18, MACC1, ECT2L, POU5F1, POU5F1B, PPARD, PIK3CB, C17orf97, LINC00511, LINC01121, SERPINE1, MIR149, MACF1, PAK1, PRKN, PCNA, MIR148A, MIR146A, PDCD1, MIR145, MIR143, MIR141, MIR139, MIR137, MIR126, LINC01194, POLDIP2, MIR106A, MIR31, MIR206, LGALS3, LINC01475, EP300-AS1, KCNH1, CRTC3-AS1, KISS1, KIT, TMEM220-AS1, C5orf66, LEP, OBI1-AS1, JUNB, LINC00446, FSBP, RTEL1-TNFRSF6B, COMMD3-BMI1, LRP6, LSAMP, EPCAM, BAALC-AS1, TMED7-TICAM2, JUND, JUN, SMAD6, IL6, H3P10, H3P23, MTCO2P12, IGFBP7, IL1A, IL1B, IL2, LINC01173, IL4, LRP1-AS, JAK2, LINC02264, IL10, CCND2-AS1, IL17A, LINC01500, IRS1, LINC01708, LINC01917, ITGB1, SMUG1, CD24, MIR21, MUC2, MST1, MIR34B, MIR34A, MIR30A, MIR29B2, MTR, MIR29B1, MTX1, MUC1, MUC5AC, MIR17HG, MIR29A, MIR27A, MIR23A, DRG1, MIR224, MIR221, MIR214, NFKB1, NHS, MSH3, MIR133B, MZF1-AS1, CRNDE, RNF19A, HOTAIR, PNKD, UCA1, MCL1, POU5F1P4, MDM2, MET, MGMT, POU5F1P3, MRC1, MIR497, MIR451A, MIR375, MMP3, MMP7, MMP9, MIR338, MIR135B, MPZ, DCBLD1, SGSM3, FAM182A, ZNRD2, USHBP1, TMTC1, CIB1, FSD1L, CXCR4, ARHGAP24, CABLES2, REEP5, AIMP2, C1orf21, B9D2, HMGA2, CD276, UGT1A7, L1TD1, NDRG1, KCNIP4, PPP1R2C, UGT1A1, REG4, EFCAB2, OR1J2, XRCC3, IL23A, TWIST1, TXN, SLC22A16, TOX2, DCTN6, TDP2, TUBA1C, SPSB2, GDE1, VDR, VEGFA, VEGFC, EZR, VIM, WNT5A, XPC, AHSA1, ACTRT3, TTC22, BICC1, GPATCH1, FAM193A, TRAP1, FRY, ACTR1B, GNG12, KLF4, NAT2, GAS5, EIF4ENIF1, ADIPOQ, GRAP2, NTN1, SCO2, DIP2B, GDF15, CLOCK, PREX1, CHPT1, SLC12A9, KIAA0355, PTTG1, NOD2, MYNN, FSD1, DHDDS, NANOG, TNKS, BAALC, BECN1, PLEKHG6, RNGTT, MUL1, ACOXL, CLEC3A, NR1I2, SPHK1, CRTC3, HERC2, MBD2, HSPB3, CFAP44, CLDN1, TRPC6, TANC1, TP73, CXCL12, RAF1, PLCH1, SPIN3, DDX53, IGF1, PCNX1, RELA, REV3L, RNF4, RETREG3, ROS1, DENND5B, CBLL2, BABAM1, S100A4, SATB1, ATXN2, CD274, CCL2, RAD51B, RAD51, RAC1, ZMIZ1-AS1, MAPK1, MAPK3, MAMSTR, MAPK8, MAP2K1, MAP2K7, PRNP, NEAT1, PSG2, HPGDS, ACAD8, PSMD9, PTGS1, TBC1D9, RTKN2, PVT1, MYO16, PDCD4, CCL20, ZNF827, TOP1, NEK10, TCF4, TBX3, SLC25A26, ZEB1, SMR3B, BTBD9, TERT, TFF3, IL17F, LILRB1, LIMA1, HSPH1, TIMP1, TIMP2, TIMP3, SEPTIN9, MFSD4B, IL33, TENM3-AS1, ADIPOR1, STAT3, STAT1, SNAI1, ZHX2, IL22, SCAF8, FOXP3, SLC2A1, CBLN2, RASSF1, RIPOR3, FSCN1, SST, TMED7, SLCO6A1, SOX2, SOX9, PTP4A3, SPP1, PPP1R21, PIGU, IGF1R, NAT1, E2F1, CYP2C9, ATM, HSP90AA1, COL4A2, FGF2, BMI1, HSPB2, CRK, DOCK3, CDC42, RUNX3, CD34, HSPB1, CYP24A1, ETS1, RUNX2, CYP2E1, MYRF, ERCC1, DNMT3B, EREG, DNMT1, CDX2, ALOX15, JAG1, EPHA2, CEACAM5, MTOR, CCK, ERCC5, DLD, COL2A1, TMEM258, CRP, ESR1, EGF, HSPA5, COL11A2, HSPA4, ACTB, HPGD, GSTT1, HRAS, CLDN7, CEACAM7, GSK3B, BAAT, GABPA, APEX1, GSTP1, BRCA1, CD68, HES1, CEACAM3, FANCC, ACE, FANCE, CHEK1, GBAP1, GAST, EZH2, FCGR3A, CASP3, FASN, FAP, COX8A, CDK1, XIAP, CASR, CD44, F3, CXCL1, CDX1, GSTM2, HFE, AREG, CDKN1B, CDKN1A, HIF1A, HMGB1, FOSB, FOS, MAPK14, HLA-A, ICAM1, ALDH1A1, EPHB4, CLU, FLT4, EPHB2, IFI27, CDK4, ALB, NR0B1, FLT1, GLI1, FOXM1, CTLA4, EPAS1, FOXO3, IFNG, CYP1A1, CDK8, CDKN2A, FHIT, FGFR4, HGF, ERBB3, ASCL2, HNF4A, ALK, ERBB2, GH1, CREB1, LCN2, SULT1A1, MIR204, CAV1, EPHX1, PSMD10, MIR125A, PPARGC1A, PRDX2, ZFAS1, UNC5C, UGT1A6, ARSA, CXCR3, MIR552, PDGFRB, MYD88, UGT1A, MIR223, AIM2, EIF4E, CDK2, DKK1, SYT1, THBS1, SKP2, LOX, MIR378A, KLF6, NTS, XIST, STAT5A, FZD7, REG1A, MIR215, PROX1, BBC3, CCAT1, MMP13, HMOX1, INSR, NOS2, TNFRSF10B, TNFRSF10A, CNR1, HDAC2, CD82, MIR32, MMP14, USP22, HLA-G, HAVCR2, SOCS1, NDRG4, DNMT3A, GORASP1, HPP1, AGT, SETD2, FMNL2, SNHG6, MYB, FSTL1, ALCAM, NF2, MSMB, MIR140, AKT2, TUG1, MIR222, DPP4, CEACAM1, MIR196B, KLRC4-KLRK1, MEG3, AGR2, CPOX, ANGPT2, ANXA5, MIR144, CCNB1, WNT2, SOX4, TMSB4X, GC, HDAC9, FOXK1, PDXP, PLAGL2, DCN, ERBB4, TNC, DKK4, PXN, MTA1, COX5A, TMPRSS13, LATS2, CD163, IL23R, EPHB3, KEAP1, SNHG1, PRKCA, MBD4, PLA2G4A, CDH13, CCN2, KLRK1, S100A9, LMLN, AR, CRYZ, SOCS3, CLDN2, SLC9A3R1, PRKAA1, IL13, ST6GAL1, KCNH2, STAT5B, FAS, AKR1A1, MIR15A, EGR1, PTPA, MIR96, HDAC1, ATF3, ST13, UVRAG, MIR486-1, ITGA5, NR1H2, AQP5, CXCL10, HMGA1, LRG1, NOS3, CDKN2B, MIR320A, NUDT1, ALOX5, ROBO1, MIR19A, CEMIP, ADAM17, CIP2A, FGFR1, ROCK1, MIR499A, SQSTM1, CYP2B6, MRE11, IL18, ANO1, RETN, TET1, OLFM4, KCNQ1OT1, SELE, NLRP3, PIK3R3, LDLR, CCR6, BIRC3, CCN1, XRCC6, LGALS4, SMARCA4, SUB1, PECAM1, PITX1, BNIP3, SLIT2, SMC1A, SPOP, SIX1, GSTA1, MIR760, FOXQ1, AXIN1, CXCR2, MIR10B, MIR142, KPNA2, BMP3, GATA6, TAZ, CCL5, MYCN, CCL21, PEBP1, IRS2, MIR22, H2AX, COPS5, ACVR2A, ANG, SCD, TRIM21, SDC2, AFAP1-AS1, KRT19, MIR608, THAS, KLK6, RARRES2, PTK2, NEDD9, CTSB, CA9, GLS, PTCH1, TMEFF2, UBE2C, PGR, CCKBR, REG3A, ANXA10, CASP2, PLG, KDM1A, PDCD1LG2, SET, SOD1, SOAT1, SGK1, HLTF, SNAI2, VANGL1, COL18A1, BSG, DDX3X, PRRT2, MIR382, XRCC6P5, UHRF1, NOTCH2, SDC1, RNASE3, TAGLN, PRKDC, MIR34C, BRD4, PRKAB1, PRKAA2, FTH1, MIR455, BLM, ADH1C, CYP19A1, PPARA, VPS51, GRN, PLCE1, PMS1, LIN28A, CST1, HSF1, PER2, BDNF, PPP1R13L, GAB2, FCGR3B, CHFR, ITGB3, TPM3, CFLAR, MIR210, ERCC4, KHDRBS1, MUC4, TP53BP1, CDKN1C, MST1R, HTC2, CLCA1, STMN1, LAG3, MME, RAD50, APOA1, ETV4, XBP1, LASP1, LEPR, BCL9, MIR185, CYP2W1, XRCC2, F2RL1, HNRNPA1, GOLPH3, VIP, MIRLET7B, IFNAR1, APAF1, TKTL1, TACSTD2, ELAVL2, BCAT1, CASC2, ADIPOR2, PIWIL1, FOSL1, NBN, MIR27B, PANDAR, ILK, B2M, THBS2, IDO1, IL1RN, MIR491, SMAD5, STC2, CXCL5, APOE, CREBBP, MIR372, NCOA3, SEMA3F, ADGRE5, KDM3A, PLCG1, SHMT1, MIR216B, SHH, MIR429, SLC1A5, ADGRE2, LOXL2, PLA2G1B, SERPINA1, MIR424, NLRP2, BACH1, DUSP4, MIR873, PGF, TRIM13, PPM1D, HLA-E, DICER1, L1CAM, AICDA, NUP62, PRL, CDK9, REN, TBPL1, RSPO3, DANCR, NOX1, RASA1, MIR622, RHBDD1, SALL4, CTSD, ZEB1-AS1, AURKB, PTBP1, RAD21, MIR532, GJA1, RAB1A, XPR1, IGFBP2, RFC1, EIF4EBP1, MIR202, POTEF, RSPO2, APRT, CD55, STK31, GRB2, DAB2IP, CYP27B1, NR1I3, CCR2, CYTOR, ANGPTL2, CACNA1G, LINC00460, KCNQ1, IQGAP1, NRP2, JAK1, SNHG14, SNHG15, PRKAR1A, ITGA4, CRY2, FABP6, MIR543, SERPINF1, MIR10A, WEE1, RUNX1, WNT1, CD36, TOP2A, TCF7, FOXF1, CHI3L1, PAEP, DCTN4, MIR301A, TNFRSF1B, GZMB, FOXC1, SRY, MIR122, TMEM8B, SMYD3, MIR216A, STAT6, MMP12, HDGF, SYK, CBS, TAC1, AKR1B1, HP, MIR181A2, HOTTIP, MIR211, MIR15B, FGF7, CDCP1, MAP3K7, HOXA9, HMGCR, STIM1, TFAM, SLC35A2, NFATC2, MAX, FLNA, FOXO1, SPINT1, GTF2H1, VTCN1, NEK2, TFF1, SNCG, ATN1, MIR23B, MIR107, ANGPTL4, HOXB13, PTPRZ1, PVR, POFUT1, TPI1, MIR574, MIR212, BGN, NOLC1, TFAP4, PER3, SOCS6, TGFB2, CTAG1B, PTPRC, ADAM28, MIR582, HDAC3, MIR200A, MIR33A, CCN6, MIR197, UCHL1, NRP1, KISS1R, PTPN13, LYPD5, ADM, BTG3, RNH1, RARA, PTPN11, TEK, HGS, RPH3AL, LPAR2, SLC16A4, ELAVL1, TFDP1, LATS1, MOK, MIR217, INPP4B, CXADR, GHR, PTGES, ARHGEF2, IL24, TCF21, MIR30B, TWIST2, TSPAN8, GEM, TLR3, CXADRP1, TFPI, HNF1A, GDNF, RIPK3, ARR3, RPP14, FADD, C12orf75, FILIP1L, PAK4, MIR28, DRD2, SIRT2, E2F4, ANXA3, CDK2AP2, MIR362, MIR483, MIR363, SHMT2, DUSP10, BMP7, PRDM2, RECK, CUL4A, FCGR2A, SPG7, MIR330, MMRN1, MIR425, MIR373, PRMT5, BAK1, SMO, DUSP2, DECR1, SLC16A1, KAT5, MIRLET7E, BAG1, MIR340, DVL2, LIN28B, IKZF1, MIR422A, FUT6, MIR100, RNF41, ECT2, TIMM8A, PDE5A, MIR494, BCL2L11, CTHRC1, SAT1, RIPK1, MIR519D, FEZF1-AS1, ESRRA, TNFSF9, KLK3, WNT3A, ATOH1, FGF13, PLK2, MIR503, MIR506, UGT2B7, AFP, VHL, BCL2L1, IGF2BP2, IFITM1, MAP2K4, MIR485, XRCC4, SSRP1, SELP, MIR132, XPA, KDM4B, PDIK1L, MIR490, NPRL2, PHLPP2, EFNA1, DAPK1, MUC12, GATA2, CYP2C19, CTAG1A, HOXA4, CXCL16, HSPA9, KIR3DL1, CEBPB, AGO2, MARCKSL1, CASC11, CRYAB, CDC73, TP53COR1, ABHD5, MYBPC3, COMETT, MIR650, MIR638, DKK3, MYH11, UGT1A10, MYOD1, ITGAV, POLB, CEACAM6, NCAM1, MAP3K8, ITGA2, LTA, FTO, RETREG1, PBK, LAMTOR2, LGALS1, ESRP1, LDHB, MCTS1, CASP7, SLCO4A1, MSI1, MSX2, GPX1, GPX2, TENM3, PON1, CMAS, MCAT, FAM172A, NR4A1, BLACAT1, SNHG16, SLCO4A1-AS1, HSD17B1, CRY1, GAEC1, LPA, CMA1, CDA, HOXD10, MIR708, CDH17, WLS, IL12B, NFKBIA, CCR7, PCDH10, MDM4, PLS3, PITX2, RMC1, PNPLA2, HOXB7, MIR1290, CMPK1, H3P8, MDK, PLAUR, HSD17B7, CDK6, ZBTB7A, CEACAM4, HK2, YBX1, CD47, CCR5, DEPTOR, NOTCH3, CBR1, MIR944, HIC1, TLR7, NT5E, SF3B6, MIR590, CSNK1E, NEUROG1, HACE1, TNFRSF11B, RAB8A, MIR130A, FRTS1, XRCC5, MTHFD2, HNRNPC, NAT10, UCP2, MIR196A2, PGPEP1, UBE2N, CISH, MIR191, MIR192, BCL3, KMT2A, GNA13, FGF4, CTCF, HOXA5, TYRO3, HOXA@, MIF, VCAM1, SPRY4-IT1, CD86, PINX1, FGA, FGF9, WNT11, WT1, BANP, MFAP1, MIR186, UPP1, CD28, UCK2, ST6GALNAC2, PTPN12, NR2C2, CD14, AHR, NR2F2, TFF2, CDHR5, CYP2R1, TOMM34, NEO1, NEDD4, MIR198, TBL1XR1, CCR1, TIA1, LINC01672, FOXE1, MIR25, RAB40B, CKAP4, TERF2, TEAD1, HLA-C, TDGF1, TDG, TRBV20OR9-2, KIF2C, MIR302A, MIR1260B, TCF12, NLK, DMBT1, AZIN1, TAP2, TAP1, HOTAIRM1, FOXD1, UGT1A8, HPSE, MSX1, MIR199A1, MIR199A2, MT1G, MIR19B1, TTK, FOXA1, TSC1, FHL2, TRPC5, MTHFD1, ALOX12, HSP90B2P, TRAF6, ZG16B, TNFAIP3, ANXA2, DNASE1, MIR205, TPT1, CKS1B, FGL2, HMGCS2, PTPN6, DHRS11, VEGFD, MUC6, MYCL, MIR219A1, UGT1A4, YWHAZ, RASSF7, CHGA, ANKRD36B, CCAR2, MTSS1, VNN1, TRIB3, TUSC7, AQP1, RSPO1, PCBP4, BCL9L, IRF2, RASSF6, GOPC, ITGA6, F2RL3, EIF3E, HNF1A-AS1, HTR1D, APLN, ITGB2, HOXA-AS2, HNRNPDL, ATP5F1A, METTL3, IL21, DLEC1, KCNN4, SIK1B, SETDB1, CES2, HSPD1, CDR1-AS, ZNF410, ERN1, MELK, EIF6, NFE2L3, PRC1, MRPL28, PDLIM7, IDH2, IDUA, ARG1, ADGRF5, CTTN, CDK5, ENG, KIDINS220, INHBB, IGFBP4, ZFYVE9, ADGRG1, IFNA13, IGF2R, LONP1, PPIG, TINCR, IL2RB, BUB3, IL4R, CXCL14, EPO, ING2, FOXK2, TP73-AS1, ARNTL, IL16, IL15, SLC16A3, AIFM1, ATG12, RHBDD2, IL7, H3P9, ROCK2, KIF22, AKT3, TFAP2E, MARCKS, TNFRSF19, CDK2AP1, DSC2, MLRL, ROBO3, SPON2, MAD2L1, TM4SF1, ABI1, LUM, ANAPC10, DUSP5, TUBB3, APBA2, F9, CNPY2, CARM1, MAP6, HDAC7, MLLT10, ZIC1, MECP2, ZNF148, HOXB8, CD40, HOXB9, ZNF654, ACVR1, MCM2, USP7, DEK, MBL2, CEACAM8, TUSC3, ADHFE1, NANOGP8, LMNA, E2F5, SIK1, PLB1, HSD17B13, HUWE1, ASIC2, UBA2, XPO5, GJC1, NUMB, SULF2, SGO1, EWSR1, KRT7, MECOM, SAV1, HDAC6, AQP8, EEF1G, MSLN, EDNRA, F7, PLA2G6, GPA33, SPARCL1, GDF11, NR0B2, AZGP1, DYRK2, PRNCR1, ROR1-AS1, DHX32, LDHA, PAG1, RPSA, LAMC2, GRHL2, IFNGR1, DDX5, MIR449A, TRIM29, B3GAT1, ATF6, MAP1LC3B, RAB3GAP1, MIR630, MIR874, GPC1, GSR, ZG16, PIM1, MIR885, OPN1LW, PIK3R1, DDX58, GPI, AJUBA, MIR20B, SHBG, MIR433, GRP, SRSF3, MAPK9, ADRB2, DUXAP9, ANTXR1, FUT8, PDPK1, SLC46A1, NDUFA13, SNCA, PDGFRA, PLAAT4, PDK1, IKZF3, BMPR2, PDK4, CASP9, SMARCA1, PUF60, RRM2B, SLIT3, KLK10, HNRNPA1P10, BAMBI, CPT1A, MIR361, MUC16, RBL2, MIR384, SLC6A4, SLC25A3, SNHG12, SNORD14C, MIR92B, SLFN11, MIR761, RPN2, MIR146B, DAP, MIR675, SIRT3, BTG1, SIRT4, MIR769, MIR495, SIRT5, PPP1R1A, SNORD14D, SOX7, TRAPPC9, DACH1, RPS18, ATG4B, RPS24, NBEAL2, HIPK2, NOB1, PHLPP1, PPBP, CCL4, CAPS, CYP3A4, CAPN2, ATF2, PLD2, MIR147B, SLC39A14, SNX10, RASSF10, IL37, SNORD14E, NR3C1, NKD2, CRIP1, CXCL11, PRRX1, TNIK, PCDH17, KLF9, NXT1, MAGT1, PODXL, CCL19, RPE65, CCL15, BTF3, MIR342, SLC25A21, SNORD35B, ARL11, SREBF1, PAX6, CTNND1, ADAMTS5, CPEB4, PTK7, NTSR1, ANIB1, AKAP10, MIR592, CSK, PBX3, GALNT6, MIR7-3, PADI2, MIR7-2, MIR324, TAS2R38, GCLM, ROR2, LINC00328, BMP1, DOT1L, MIR598, MIR551B, CXCR5, COMT, RAB27A, MIR1246, GCLC, GSDME, OGN, FURIN, PTX3, PIM3, STC1, MIR600, ADAR, STEAP1, WWTR1, PSMB8, MIR7-1, FOXD3, MIR339, COL1A1, GUCA2A, MIR335, SIRT7, PTPN3, FOXD2-AS1, SYP, MIR328, SPINK1, PTPN1, PSMA7, SP1, PART1, PTPRG, FRZB, VCAN, EDNRB, MAP3K21, SRPX2, GSTO1, SELENOS, CTNNA1, RANBP9, KRT23, ITGBL1, ATP6V0C, LCS1, BCAP31, RGCC, EGFL6, LYPD4, MTA3, EMX2, DPPA2, EML4, CENPA, BEX2, IL17C, CDC6, CDC25C, PDCD6IP, SUZ12, FGF21, MAP4K4, AXL, NAV2, ABCC10, BZW2, APOM, DCP1A, SPZ1, ALX4, MIIP, RASGRP1, CYP3A5, CYP7A1, TGFBRAP1, ATR, EEF1A1, TSPAN1, ARL4C, ROPN1L, PROK2, WASF2, ATP6AP2, ENO1, ZNF518B, MIB1, TDP1, FAM126A, CRMP1, EEF1B2P2, AVP, EFNB2, CAPNS1, EBI3, RABGEF1, RCC2, FBXO8, ARHGEF12, RPPH1, BHLHE22, CA8, MVP, ATP5PF, ANKS1B, FANCM, PARP6, SND1, ARNTL2, ORAI1, BAG3, KIF4A, IL17RA, TOX, EIF4A3, DLGAP5, CX3CR1, PNO1, PLA2G15, RNF40, ENAH, FOXP1, CALCR, ECRG4, NLRC5, MED19, CDKN3, MINDY4, CTSL, ASIP, CD177, CSF1, KLK5, PAK5, RGS6, CYTIP, IL17RC, NLRC3, EIF5A, CTSK, SPDEF, ASNS, PLXDC1, CDH10, LINC00467, ASPA, RAB3D, ASS1, PRSS55, TIGAR, CBX8, SPATA2, EIF2AK3, PROK1, LPAR3, BCCIP, CNRIP1, CA2, MEGF8, RPRD1B, KIF14, MPEG1, ARHGAP25, ABHD11-AS1, SNHG7, NDRG3, LY6G6D, SBDSP1, SNORD14B, CDK7, ZNF281, CDC27, HRH4, SEMA6A, CASP8AP2, EIF5A2, UBAP2L, NUAK1, PATZ1, TRIM14, ADAMTS3, ADAMTS4, CD2AP, FAM30A, MAEL, HAMP, RAD18, POT1, SPHK2, ARHGDIB, SLC2A4RG, FITM1, PDSS1, DKK2, RAB22A, MUC13, NUPR1, SLC5A8, TMPRSS4, ESRP2, DROSHA, SMOC2, CASP6, SRCIN1, ASPN, GGCT, CD3EAP, CLCA4, MGLL, AZIN2, TRPM8, POLR1D, GAL, MLXIPL, CLDN4, TMEM54, BTG4, BUD23, KLF8, SARDH, KLF12, CKS2, CD247, CORO7, LYVE1, SH3GLB1, MECR, DES, VASH1, CLDN3, PSMD14, SETD7, CMPK2, MAPRE1, TESC, FAM83D, CD209, USP39, TXNDC5, POLQ, FAN1, PLK4, IQGAP3, KLF17, YTHDF1, BCR, RUFY3, PLA2G3, STK33, CPT2, TP53INP1, MS4A1, BCYRN1, ASAP1, CAVIN3, EGFL8, TRAT1, PAF1, CAT, DDX56, ING3, COL6A3, HAND2-AS1, COPS6, SIX4, MAP3K20, CDC37, MAPRE2, NAA40, PEAK1, UBR5, TRAPPC4, IMMT, POLK, ISYNA1, PARD3B, NHEJ1, MFSD13A, LINC00472, WDR76, VTA1, TOPBP1, TRIM31, ESM1, SIRT6, COL1A2, SHC3, P2RY13, BVES, EHMT2, GALNT14, FAT4, IRAK3, CCNG2, CPSF4, JTB, FZD10, BRD8, CLN3, PABPC1L, SLC31A1, EGFL7, CORO1A, H4-16, LMTK3, SUV39H2, PALB2, TREM1, SLC25A22, RALBP1, RERGL, WNT10A, COL11A1, MYSM1, MARCHF1, BRS3, USP47, CKS1BP7, CD63, RASSF5, RACK1, PIAS3, ST3GAL6, ADGRB1, CARD9, DUSP1, ZC3H13, IL17RB, DSC3, TRPM6, VAV3, KDM4C, BATF3, MAD2L2, PDLIM2, CRTAP, TRPV6, RHBDF1, ZNF331, UPRT, AMER1, RNF183, IMPACT, SPART, NOD1, BTK, E2F7, SAPCD2, TRIB1, UCN2, CCDC88A, STARD13, SPRY2, C3, CTSC, MOAP1, SIGMAR1, FMNL3, E2F3, ITLN1, DDX27, PIWIL4, CD70, VSIR, TLR6, CGA, FERMT1, CRIP2, OTUB1, BAD, TSPO, STYK1, NEBL, DOCK2, ATG7, A1CF, OBP2A, TLR10, PDLIM5, ADAMTS12, TRIM3, DCLRE1C, CSMD1, VMP1, TXNIP, NDST4, PDPN, ANAPC1, NUCKS1, PRMT6, CHKA, SMURF2, IGF2BP1, GALM, CHRM3, RPIA, CELF2, CXCR6, SLC52A1, ATG16L1, BCL2L2, MOB1A, HNRNPLL, BST2, PRSS50, UBD, CRHR2, USP33, CD40LG, CDCA3, TCF7L1, BCHE, ANP32B, LINC00312, HTATIP2, SLC25A24, SCGN, PSPC1, SPEN, CXCL13, DNER, MRGBP, ENTPD1, SLC26A3, CD38, SLC34A2, PSAT1, DDB2, CD8A, APOD, RBP4, MIR367, SRI, FUS, MIR148B, MIR3666, MIR302C, SPTAN1, HLA-DRB1, SPOCK1, MIR326, SPN, MIR331, FUT4, NEU1, SNX1, G6PD, TLE5, SSAV1, LINC01133, MIR99B, CBR3-AS1, PANTR1, MIR98, NR5A2, MIR99A, STK3, STAT4, SSTR2, EIF2AK4, LUCAT1, NCL, MIAT, SSX2, SSTR4, NEUROD1, SMARCC1, AGTR1, SMARCB1, HNP1, GAS1, MIR452, MIR409, MIR410, NONO, MIR488, MIR489, GATA1, CX3CL1, MIR511, CCL24, CCL22, CCN3, CCL11, MIR18B, PGM5-AS1, H4C15, MIR3163, ACYP2, SLC22A5, SLC22A4, NFIL3, MIR466, NGF, MIR383, SIM2, SLC5A5, MIR423, DUXAP8, SLC2A4, DUXAP10, SKI, TAPBP, MIR30D, CCL3, LINC00707, WNT3, VWF, MIR4500, AMFR, MIR152, MIR154, HNRNPK, HNRNPD, MIR181C, ALPP, ALPI, MIR187, USP4, FOXA2, UBE2V1, FES, ANPEP, MIR136, CNBP, FBLN1, FBN1, MIR1-1, MAP3K11, TRIM25, FCGRT, ZFX, MMP16, ZFP36, HOXA6, YY1, MMP8, ANXA1, MIR4478, UBA52, HMMR, MYO6, FGL1, PPA1, FLT3, TFCP2, MIR29C, TFAP2A, TERF1, TERC, TEAD4, PPP1R11, TCP1, TCN1, FOLR2, FOSL2, MIR30C1, MIR30C2, TGFA, MVD, MIR296, TP53BP2, MIR199B, TTF1, TRAF1, HSP90B1, CRISP2, HMGB3, MUC3A, TGFBI, FOXF2, NKX2-1, FOXC2, TRIM37, THPO, AHSG, CCL7, SCTR, PTK2B, PIN1, PTN, PGK1, PGM5, PHF2, GLP1R, PTH, MIR597, PTGER4, PTGER2, GLUD1, PSPH, GMDS, PSMD4, MIR939, SERPINE2, PGC, MIR579, MIR577, GHSR, PDE4B, PDE4D, MIR411, RAD52, RAD51C, TRAP, RAB27B, GLI2, GJB2, PF4, GLB1, NECTIN1, KIR2DS2, PTPRK, MIR603, MIR942, PDE4A, MIR605, UTS2R, MIR663A, FXYD3, PLOD1, MUC5B, GPR42, PLXNA1, ARHGAP35, PPP2R2A, GGTLC3, MIR766, GGT2, POMC, HULC, GGTLC4P, PLD1, PLCL1, MAP2K2, MIR625, GNA12, MIR374B, HTRA1, KLK7, RELN, CXCL3, GOLGA2, MAP2K5, LGMN, MIR627, MIR629, GOT2, PRODH, MIR744, RAP1A, RAP1GAP, GBP1, ADRA1A, MIR520G, GCG, RRM1, RPS27, RPS20, RPS15A, MIR505, MIR508, HCC, RPLP0, RPL29, MIR1827, RPL22, OPA1, RPL10, RXRA, SORT1, NUP88, MIR181D, NPC1, NPM1, SCN1A, NPR3, HGD, ADRA2B, MIR498, DDR2, NRL, NTRK2, NTRK3, S100A11, ROR1, S100A10, LEF1-AS1, PURPL, PCYT1A, RP1, RCN2, MIR539, PAM, MIR544A, PAWR, RBM3, RBBP6, MIR487B, RB1, PCCA, LINC01630, RASGRF1, RARG, GGT1, GUCA2B, REG1B, GFI1, SNHG20, MIR1229, RORA, SNORD15A, GDF1, MIR1297, RNF6, MIR1258, OTX1, UPF1, RNASEL, NORAD, GGTLC5P, P4HB, REST, MIR1914, FZD5, MED1, BIRC2, CLMAT3, INPPL1, CUL1, OGT, PRMT1, ACP3, FOXP4-AS1, CBX4, PKMYT1, AP3B1, LINC00265, IAPP, LIPC, LIMK2, IREB2, PWAR4, LIMK1, KHSRP, ANGPTL1, LIG4, IRF1, IRF3, GPRC5A, CADPS, HCA1, CYP4F3, LTBP2, H4C14, H4C3, H4C8, HOXD8, H4C2, H4C5, IL1RL1, COX7A2L, ACTN4, H4C13, LOC110806263, MAFG, SNHG5, ILF3, HPS1, MAD1L1, CBFA2T2, ING1, ASPM, ABL1, LTBP4, TMEM189-UBE2V1, TMEM189, WSPAR, HSPA1A, H4C12, IRF7, JAG2, MIR6869, TRIM59, CASC19, FUBP1, RAB11A, ARHGEF7, TNFRSF6B, TRIM24, TPTEP2-CSNK1E, ANOS1, CDKL1, MIR6803, KIR2DS1, FGF18, SOCS2, CCN5, CBSL, DLX6-AS1, KSR1, GHET1, HSP90AB1, ACAT1, ESRRB, GACAT3, CCNA1, LBP, FBXL19-AS1, CISD3, EXTL3, LCT, DYNLL1, LCP1, EIF3G, ITGA1, PEA15, ACLY, HSPA1B, ITGB4, ITGA3, ITGA7, ITGAE, LAMB1, BTRC, HSPA2, ITGAL, CERNA3, PERCC1, ADGRE4P, H4C11, FASLG, H4C6, IFIT2, USP9X, CXCR1, FABP2, KITLG, HOXB6, MGP, MEIS2, H4C9, FABP4, H3P47, IGFBP5, GTF2H5, NNT-AS1, GCNT3, CIITA, SHOC2, HOXD3, BCAR4, MAPKAPK2, FZD1, IL6R, MEIS1, RICTOR, SLC7A5, MAP3K1, IHH, MEN1, MEP1A, IKBKB, H4C4, HAGLR, MFGE8, NRIP1, RHOA, COPS2, MELTF, ME1, ID1, APBA1, IL9, EPHA8, CCDC6, MBNL1, MIRLET7C, IL32, FEZF1, MATN1, MAS1, MIP, C2orf68, SNHG17, EPHB1, SLC16A7, ACSL1, OSMR, LINC00365, AANAT, H4C1, IFNB1, FZD6, CDR3, CXCL9, IL11, FAAH, MC4R, NCOA4, MIR1182, DYNLRB1, NUF2, MIR1288, CASP1, CBFB, MIR1307, H3P28, MIR1275, MIR1296, NUBPL, EPG5, ADD1, TP53INP2, IBD27, FAM3D-AS1, ZGLP1, MIR1295A, MIR298, JAM3, NRIP2, ADARB1, NCOA5, SCUBE2, SNORD12B, MED28, MIR1256, ULBP2, MIR1249, EFHD1, CXXC4, CAV2, MIR1247, ACADM, WDR82, RGPD2, MUC3B, SNORD126, ZNF398, ADD2, MIR454, CDK5RAP3, H19-ICR, PTBP2, MIR1976, SNHG8, NLRC4, CAST, CASC21, USP28, SLC25A25-AS1, OLA1P2, MIR365B, CASP5, CDH4, XIAP-AS1, ALPK1, GDPD5, LOC112543491, LINC02605, STIM2, HM13, CDH15, MIR877, CLPTM1L, WNT5B, FAM83H-AS1, VPS9D1-AS1, CASP4, CDH12, ACTL8, TRIM8, MIR938, MIR935, MNS16A, NETO2, LOC111365171, WDR48, MIR940, MIR889, TRMT9B, LOC110599580, NECTIN4, LOC110283621, MIR297, SNORD138, SLAIN2, KTN1-AS1, MIPEPP2, LOC100288966, CALCOCO1, DIP, ACAA1, AFA1, RAB1B, CCEPR, MIR888, QTRT1, LINC02418, LNPK, CHD8, ABCF1, ACACB, SEMA4G, MIR450B, METTL14, KIF18A, CASP10, MYH7B, TAOK1, EP400, LUADT1, ABO, ADCY3, FOXO6, DDHD1, NR4A1AS, LOC106128905, ZBTB2, ADCY1, UXT-AS1, MAP3K14-AS1, RNF146, MIR875, SPRY4, H3P13, GINS3, IATPR, RUNX1-IT1, PCAT6, LINC01234, ST3GAL6-AS1, DSCAM-AS1, LIFR-AS1, TCTN1, LINC00400, OCLN, SAP130, DDX11-AS1, LINC01354, CD58, NOC3L, GIHCG, DLEU7-AS1, FALEC, TTN-AS1, TTPAL, MELTF-AS1, MMP28, DHX58, CERS4, CCT6A, OVOL2, LINC01503, IQCJ-SCHIP1, FN3K, CA3-AS1, GAPLINC, SH3PXD2A-AS1, FOXD3-AS1, MAGI2-AS3, RIOX1, TINAGL1, DNAH17-AS1, LINC00649, STEAP4, LINC00958, CCT, LINC-ROR, BANCR, BEND5, IQCJ-SCHIP1-AS1, MCPH1, STARD13-AS, ADAMTS9-AS2, MAPKAP1, ITIH4-AS1, PSC, ACVRL1, POU6F2-AS2, RTN4R, PINK1, CD80, MIR4487, MRPL41, MIR4717, MIR4666A, MIR2467, HEIH, CD33, DCLRE1B, LINC00659, MIR4775, MIR4492, SIGLEC6, USP46, MIR378F, MIR4658, ACY1, BCL2L2-PABPN1, UBE2Z, HIF1A-AS1, MEG8, RNF26, DSCC1, MIR5191, ENTPD5, METTL16, CT47A12, SECISBP2, CUEDC2, HIF1A-AS2, VIM-AS1, SOX21-AS1, CAHM, RPL17-C18orf32, TMX2-CTNND1, CD27, CDH24, POC1B-GALNT4, ZSCAN18, MIR3150B, SLC39A8, E2F8, AK6, CDC25A, MIR6852, NCRUPAR, SEPTIN7, MIR6787, WWC2, RMI1, MIR663B, MIR6165, MYMX, SATB1-AS1, STN1, MIR4474, APOBEC3G, BRCA3, RINT1, ACHE, BOK-AS1, ZBED6, ADAM8, ADAM10, AFAP1, CDC25B, ZNF606, MIR1266, CHD9, MIR6868, HKDC1, CWH43, MIR1179, ASRGL1, MIR6778, MIR1236, SERPINH1, PTGES2, MIR6716, MIR8075, MIR1910, ZNF703, NTN4, FTX, MIR8064, MIR7702, CDC5L, ACR, CCNE1, MIR3679, ACOX1, ADA, CAMKMT, EHMT1, CCNC, PARVG, MIR3621, MIR3678, MIR642B, CD81, FKBP10, EZR-AS1, SHCBP1, CARF, MIR3940, LSINCT5, ACP5, OPN1MW3, ELMO3, NPY4R2, PRDM14, CASC9, MIR4282, MIR4261, SMAP1, HEATR6, MIR4319, KRIT1, MIR3191, SLC17A9, ELFN1-AS1, CNTD2, MIR514B, DOK3, CCNA2, GALNT11, XPNPEP3, CYBRD1, ACO2, MAP9, MIR4316, MIR4260, NEK11, CD2, SPDYA, OIP5-AS1, STK35, DACT3, ZNF582-AS1, AVPR2, CCBE1, NOTUM, ACF, C10orf143, KMT5A, ZNF785, TMED6, IL34, CMTM2, CMTM4, ZFP90, LINC00052, LINC00355, KRT80, CCL4L1, MUC15, XRRA1, DPP10-AS1, BEND4, PSMA8, FLT1P1, EIF5AL1, MIR100HG, GSEC, ZNF583, USP17L2, MIA3, ZFP57, STING1, POTEA, APP, ASPRV1, APOC3, RFPL4A, GPBAR1, ATP6V0A1, ARL6IP6, CYCSP51, CDKL4, ACTBL2, SATB2-AS1, ZNF829, ZDHHC8P1, APOC1, ATRX, GEN1, APOBEC1, LINC00654, RTN4RL2, ZACN, APOA4, BNIPL, DND1, PTPRQ, ST20, FENDRR, ANKLE1, ANXA13, AFAP1L1, PPARGC1B, IL31RA, AASDH, MIR133A2, MIR134, CPEB2, TMEM207, ANGPT1, BCL6, ANCR, AMT, RBM45, BIN1, AMELX, MIR153-1, AMCN, AMBP, ALX3, DIS3L2, MIR184, ZNF280A, TNFRSF17, PROKR2, SNHG11, NAXE, MIR188, GPR151, MIR125B1, MIR124-1, CASC15, ANXA11, ROMO1, LINC00668, LINC00909, ADAMTS14, ANXA7, MACROD2, CTCFL, GCNT7, ANXA6, S100A16, MUC17, ACTRT1, MUCL3, HOXB-AS3, GAB3, FOXR2, PASD1, TAF1L, MIRLET7D, VPS37A, MIRLET7I, MIR103A2, ANXA4, CLDN23, KLF14, BTLA, GADL1, MAP3K20-AS1, ATP6V1A, SGMS1, ENKUR, MRGPRX1, COPD, HIPK1, STEAP2, BEST4, SELENOH, TIGIT, ARRB1, TRIM23, TRIM65, SBF2-AS1, ARSB, FBXO45, RPL22L1, CRYGEP, ART1, TXLNA, DAND5, H19, ASAH1, B4GALNT3, IMMP1L, B3GNT6, PGAM5, EWSAT1, ARF6, UNC5B, LY6G6F, MCM9, RHOB, RHOC, GPRC6A, LNX2, IL27, TMEM9, FNDC5, CERS6, PRR15, VSTM2A, YTHDF3, THSD7A, LPCAT4, RHOH, HOXA11-AS, LINC00957, ARHGDIA, CT47A11, BCL6B, HNRNPUL2, JMJD1C, RNF152, HEPACAM, CBY2, ARHGAP30, TAB3, PGP, ADAMTS16, ASL, ZNF746, SLC35G1, SCARA5, NKX2-3, FIRRE, SBSPON, ATP2A3, EIF4E3, ESCO2, CDCA2, PEBP4, POTED, AGR3, ATP2B4, GRASP, SNAI3, CHSY3, TAS2R64P, ATP5F1B, SHISA3, PAQR3, ANO9, ATP5F1C, FFAR4, VSX2, JMJD8, ARMH1, FAM83H, DNAAF4, LINC00858, SLC9A9, CAVIN1, FAM27E5, AQP9, AQP6, DACT2, MIER3, SGMS2, ZFP82, ADGRA3, NWD1, DNAJB8, UBXN2A, FAM171B, LINC00996, OXER1, ATD, EPHA6, HFM1, SASS6, CITED4, CCDC66, RPL34-AS1, RGMB, ATF4, LINC00174, FSIP1, MIR190A, ALOX15B, SPANXA2, IL1F10, MIR545, TMEM25, RIOX2, TRIM52, MIR503HG, SFT2D3, MIR542, POLR2J4, SCARNA2, C19orf48, LINC00488, SNORA80E, MIR33B, CACNA1F, DDR1, MIR421, LNX1, MIR567, GPT2, MYO18B, PPP1R9B, HINT2, MIR568, ACCS, MIR572, SLC25A20, GLYR1, CA7, ATOH8, UTP4, MYLK2, TSPYL5, MIR484, TMEM75, ZIC5, ADRA1B, GAS8-AS1, NKD1, NR2F2-AS1, PHGR1, H3P16, EIF3J-DT, MNX1-AS1, LGALS12, SEMA3D, COL27A1, WFDC21P, TPTE2P1, CA4, LYPD8, NUDCD1, LINC00273, ADK, CT47A7, CA6, NME1-NME2, MASTL, MFSD14B, MIR584, HSPB6, AFAP1L2, KAT8, CALM3, CEP78, ADH7, USP44, CAMK2B, SSX2B, CT47A10, WDR54, CAMP, CT47A9, CT47A8, CT47A6, CAPG, CAPN1, CT47A5, CT47A4, CT47A3, CT47A2, CT47A1, DEFA1B, ANGPTL6, OPN1MW2, MEMO1P1, SETDB2, ATG10, CRISPLD2, MIR647, MIR646, PPP1R1B, MIR601, MIR585, MIR587, MAK16, MIR595, MRPL43, ACSS1, HOPX, ACRBP, CALB2, BRSK1, CALCA, RAB11FIP4, COG8, MIR637, MIEN1, PHF6, CALD1, CALM1, TCHP, FLYWCH1, MAF1, MIR612, MIR618, CALM2, LOXL4, RNASEH2C, SNORA21, MPPED2, MIR502, SSH2, AGAP3, AGRP, SNAP47, MIR95, BIK, PHACTR3, BATF2, ALPK2, LINC01555, ARHGEF25, PRKCQ-AS1, TRIM67, MAP1LC3C, FBXO32, SORCS1, VN1R17P, CSMD2, GPR166P, PKD1L2, AEBP1, MIR155HG, TIRAP, BMPR1B, MAL2, MIR323A, LACTB, TXNRD3, MIR30E, TADA1, MRGPRX3, C14orf28, TNFAIP8L1, MIR196A1, ZNF543, GJD3, SPNS2, BDKRB1, CCDC43, ALDOB, LRRC28, ALDH1A3, ALDH1B1, DEGS2, MIR24-1, MRGPRX4, MIR24-2, MIR26A1, SPIC, AHCY, FAM186A, BID, SLC36A4, GSTO2, MMP21, BORCS5, APLNR, GALNT15, MIR337, AP2A1, CDCA5, MIR519C, TIMD4, BTF3P11, MIR492, MIR493, MIR496, ISX, TRIM47, CCDC34, MIR193B, C1R, MIR520E, LINC00473, BMF, MTG1, IFT20, DCAF15, MIR519B, UNC5A, MIR520B, PHLDB2, MIR524, NAV3, MIR522, MIR500A, MIR501, SCIN, BTC, SPECC1, NACC1, TRIM72, MIR345, TP53RK, KRT71, MIR369, MIR370, SAT2, MIR374A, POTEE, SUMO1P3, EGLN3, EGLN2, BOK, CHAC2, MAFG-DT, ADRB3, HTRA3, MIR376A1, TTYH2, FOXP2, MIR4435-2HG, CKMT1A, MIR31HG, MIR448, NAPRT, UBE2Q2, MIR431, RPTOR, DGKB, MTUS1, SULT2A1, SUV39H1, TACR2, TACR1, TAF1, TAT, FPR2, TBL1X, TBX2, TBX5, FPR1, FOLR1, FMO5, FMO3, TDGF1P3, TFAP2C, FLT3LG, TGFB1I1, TGFB3, FLII, SUPT4H1, STYX, THBS4, STXBP1, SREBF2, SRPK1, SSBP1, ITPRID2, SSR1, ST8, ST14, FUCA1, STAT2, FTL, STATH, STAU1, SULT1E1, NEK4, STK4, CENPI, AURKC, STRN, STX1A, THBD, THM, FUT1, TPTE, TRAF5, CCT3, TRIO, TRIP6, TSC2, TTR, TULP3, FGR, TNFSF4, TNFRSF4, TYK2, TYRP1, UBE2I, UCN, UCP1, UGT2B15, UGT2B17, FGF12, UQCRB, TRC-GCA24-1, TPO, THOP1, TPM2, TIE1, FOXJ1, FOXL1, TLE1, TLE4, TLR1, FOXD4, TLR5, TM4SF4, CLEC3B, TNFRSF1A, TNNI3, TNNT1, TOP2B, FKBP3, TPBG, TPD52, TPH1, TPM1, SQLE, SPTBN1, RPS2, SERPINB3, SCN10A, SRL, CCL1, CXCL6, SDHB, SDHC, SDHD, SEMG1, GAS8, SFPQ, GAS6, GAS2, LRRC32, SRSF1, GALNT3, GALNT1, SH3GL3, SHC1, SI, SCN7A, ATXN7, ST3GAL4, GBP2, RPS6, RPS6KA3, GCNT1, RPS6KB2, RPS7, RPS27A, RRAS, RRBP1, RTN1, RYR3, S100A1, GCDH, S100A6, S100B, SAA1, SAA2, SAI1, SALL1, SBF1, SIAH1, ST8SIA1, SPTA1, SLC18A1, SLC25A1, GABBR1, GAB1, SLC22A2, SLIT1, SLPI, SMS, SNCB, SNRPA1, FUT7, SOS1, FUT5, SOX5, SOX10, SOX15, SOX12, FUT3, SPP2, FUT2, GABRA1, SLC14A1, SIPA1, SLC12A3, GALR1, SLC1A1, SLC1A2, SLC1A3, GALK1, GAGE7, SLC2A3, SLC3A2, SLC4A1, SLC5A2, GAD2, SLC6A2, SLC6A6, SLC6A8, SLC6A12, SLC7A1, SLC7A2, SLC8A1, SLC10A2, UQCRC1, FGF10, UQCRFS1, NCOA1, EIF3D, EIF3J, SLC4A4, ETV5, HYAL2, GALNT4, B4GALT3, B4GALT2, B3GALT4, SNX3, EED, ETS2, RNMT, GPAA1, HRK, TNFSF14, ADAM23, ADAM15, PABPC4, EIF3A, EYA1, TNFRSF14, AOC3, GAS7, DGKE, CST7, YBX3, BARX2, APOL1, F2R, F2, CDK10, DEGS1, DENR, MKNK1, CASK, AKR7A2, PLA2G4C, EYA2, JRK, RTCA, EIF4EBP3, CD164, TNFRSF11A, UROD, NR2F6, EPS8, TAF1C, TAF1B, TM4SF5, PKD2L1, SPAG9, ARTN, AIP, SYT7, CLDN12, CLDN8, SART1, USP14, USP2, FCGR2C, RGN, MTMR7, NMI, INA, NOL3, WASL, IL18R1, USP13, BANF1, CCN4, DLEU2, TSC22D1, KAT2B, AKAP4, ASAP2, ERG, IER3, SYNJ2, ERCC3, EIF2S2, CPNE3, MTMR3, PRPF4B, SGCE, RTL8C, WASF1, H3C7, MMP23A, MMP23B, ENC1, WNT6, XBP1P1, XDH, XK, GPC4, XPO1, FECH, FDXR, YWHAE, ZAP70, ZIC2, ZBTB16, FCGR2B, FBP1, ZYX, MEMO1, SCLC1, MAPKAPK3, BPTF, SLC39A7, WNT2B, FGB, TPST1, NSD2, USF2, USP1, UTRN, VASP, VCL, VCP, VDAC1, FGF8, VEGFB, VIL1, FGF3, VIPR1, VLDLR, TRPV1, VSNL1, VTN, WIPF1, EIF4H, LAT2, NTT, FAH, FZD3, ST7, F13A1, H3C1, H3C4, H3C3, H3C6, H3C11, H3C8, H3C12, H3C10, H3C2, PLA2G10, UXT, ANXA9, SNHG3, STK24, RASAL1, CUL4B, CUL3, CUL2, FZD8, EOMES, CDC45, GPR68, PSCA, FANCD2, PDHX, MFAP5, USP5, FXR1, YEATS4, FANCA, TCL1A, CDC7, SLC14A2, FABP5, MIA, TAM, PUDP, USP11, NAA10, FABP1, RPS4X, RPL27A, INTS2, MNAT1, MPG, MPL, HNRNPL, MPP3, CITED1, SLC29A2, HNMT, MSN, ONECUT1, MT1X, ND1, ND2, MTTP, TRNF, HMGB2, MUC7, MMUT, MXI1, GADD45B, CD200, MN1, MYH9, MMP19, HOXD@, MCM4, MCM7, MDFI, MEF2A, MEF2C, MEF2D, MFAP3, SCGB2A1, MGST1, MICB, MID1, HOXB@, HPCA, MLN, FOXO4, HOXA10, HOXA2, MMP15, MYH6, MYO1A, MARS1, HLA-DOA, MNX1, NFYB, NGFR, NINJ2, NIT1, NM, HK1, NME3, NME4, NNMT, HINT1, CNOT3, NPY, NRG1, NPAS2, NPTX1, NPTX2, NRDC, HELLS, NFIB, NFATC3, MYO5A, HLA-DQB1, MYO5B, MYO10, PPP1R12A, PPP1R12B, HNRNPM, NAP1L1, NAP1L2, NARS1, NUBP1, NCF4, NCK1, NDN, NDUFA4, NDUFB2, HLA-DRB4, SEPTIN2, NEDD8, NELL2, HLA-DRA, MAT1A, MARK1, RPL19, IRAK2, IRF5, IRF6, HYAL1, ITGA2B, ITGAM, ITGAX, ITGB6, ITGB8, ITK, ITPR3, IVD, HTN3, JAK3, HSPG2, KCNA3, KCNA5, HSP90AA2P, KCNMA1, KIF2A, IRF4, PDX1, KIR3DL2, INSRR, IGFBP1, IGFBP6, IGKC, RBPJ, IL2RG, ID4, MRPL58, IL7R, ICA1, IL10RA, IL12A, IL13RA2, IBSP, IARS1, IMPDH2, INCENP, INHBA, INPP1, INS, KIR2DL2, KLK1, MANBA, HSD17B4, HSD17B2, LLGL1, LMO1, LMO2, LPP, HSD11B2, HSD11B1, LRP5, LTA4H, BCAM, LYN, HRH2, MXD1, SMAD1, HRG, HPX, HOXD13, MAGEA3, MAL, LIPE, HSF4, KLRC1, LIF, KNG1, KIF11, KPNB1, KPNA3, IPO5, KRT5, KRT6B, KRT17, KRT18, LAMA2, LAMA3, HSPA1L, LAMP1, LBR, LCK, LECT2, DNAJA1, LGALS3BP, LGALS9, NUCB2, NUP98, OAS2, PRSS2, PSEN2, GNAT1, PYY, PSMC2, GML, PSMD7, PTGDR, PTGDS, PTGIS, PTHLH, PTMA, PTPN4, PTPN9, PTPN14, PTPRA, PTPRB, GLO1, PTPRH, PTPRO, PSD, PRS, PTS, GOT1, PPP2R2B, PPP2R5E, PPP3CA, PPP4C, PPY, NPY4R, PRCP, SRGN, PRKACB, FFAR1, GPR31, GPER1, PRLHR, MAPK7, GPR4, MAPK13, MAP2K3, DNAJC3, GP2, PTPRR, GPC3, OCA2, RGS2, RHO, RING1, RNASE1, BRD2, MSTN, RNPEP, ROBO2, RORB, RORC, OPN1MW, RPGR, RPA1, RPA3, GCNT2, RPL7A, RPL9, RPL11, RPL17, RPL18, RGS16, RGS1, PYCR1, RFX5, RAB5A, GIP, RAD51D, B4GALT1, RAN, RANBP2, RAP1B, RAP2B, RASA2, GFRA3, ARID4A, RBBP8, GFRA2, GFRA1, RENBP, GFAP, RFC3, RFC4, TRIM27, GRK6, PPP2CA, PPP1R8, PCK1, PCNT, PDC, PDE2A, PDE7A, PDGFA, ENPP1, GSTT2, PENK, PFDN1, PFDN4, ATP8B1, PFKFB3, PFN1, PFN2, CDK14, PGAM1, GSTA4, PHB, PHKB, GUSB, PCCB, PPP1R3C, PCBP1, HAS2, OGDH, HADHB, OLR1, OPRD1, SLC22A18, ORM2, H3-3B, OTC, CLDN11, H3-3A, P2RX5, H2AZ1, P2RY6, PA2G4, PRDX1, SERPINB2, GYPA, PAX3, PI3, SERPINA4, GSTA2, SERPINB9, SEPTIN4, GRB14, POLG, POLH, POLR2A, POLR2B, POLR2F, PON3, POU2F2, GPX3, PPAT, CTSA, PPIA, PPIB, PPL, PPM1A, GPT, PPP1CA, PPP1R3A, GRIN2B, PML, PMAIP1, PKLR, SERPINI1, PIGF, PIK3C3, GSPT1, PIK3R2, PIP, PKD1, PKD2, GRPR, PLXNA2, PDIA3, PLAGL1, PLAT, PLCD1, GRM2, FXYD1, PLOD2, PLSCR1, PRPF4, STX2, PDCD5, CXXC1, SOCS7, CTAG2, STOML2, KLK12, NOX4, PNPLA8, TAS2R16, TAS2R14, F11R, CPM, BHD, ASCC1, DERL2, ISOC1, DESI2, RPS27L, MEMO1, APIP, LACTB2, CR1, SOX8, ASB3, SPANXA1, CPS1-IT1, C1GALT1C1, TMEM208, BRD7, CTNNA3, RACGAP1, CARD10, PARVB, CYP2S1, ZDHHC1, GRHL1, CNOT7, EEF2K, HILPDA, NRBP1, CRAT, BICRA, ERO1A, TBX21, SAR1B, IRAK4, KLF15, SNX9, NAT8B, NELFCD, DTL, NCKIPSD, ANAPC11, RAB14, ACKR4, CNR2, CYRIB, CNN3, PLK3, RASD1, ZFR, ASB4, TPPP3, CPSF3, CNGB1, CYB5R1, RAB23, MAGEC2, AMOTL2, INSIG2, COMMD10, CKLF, CPA4, COL12A1, IGF2-AS, GLTP, PIPOX, KLF3, MAPKAPK5-AS1, GOLM1, CD320, BPIFA1, KCNK9, ARMCX1, TLR8, MEX3C, COL10A1, THEM6, UCHL5, COL5A2, ATAD2, MACROD1, SLC28A2, TMEM158, CTNNA1P1, ADGRA2, SH2B1, TSKU, TENM4, L3MBTL1, CSTB, RAB11FIP5, APPL1, CST4, CHTOP, CST3, LRIT1, LDLRAP1, CSRP1, CSNK2A1, INTS1, DGCR5, FBXL3, SAMHD1, TRIM58, PHGDH, LETMD1, CADM1, CTSZ, ARHGAP8, CTSS, TFIP11, FJX1, CBY1, CFAP45, CIZ1, GCA, LMOD1, TNFAIP8, ARIH1, MTO1, ASF1A, MOB4, PARM1, BRMS1, SUMF2, ARL5A, HIBCH, DBNL, PALD1, GLS2, SIGLEC9, IL17B, GPR162, SPINK4, RBMS3, CRX, TNRC6A, RPS6KA6, UBE2S, POLL, TMEM97, DESI1, MAT2B, SLCO3A1, IGHD1-14, DLL1, TRIB2, GIT1, NAAA, CRYGC, VPS33B, CACYBP, CSNK1D, MYCBP, HEYL, RGS17, MYEOV, CSN2, CKAP2, OR7C1, HAVCR1, SNORD44, SNORD33, RNU6-6P, HBP1, PABPC1, CSF3, LAT, PELP1, ANKRD1, NSG1, CAB39, UPB1, GHRL, OGDHL, CETN1, CES1, CENPF, ST6GALNAC1, FOXJ2, ARHGAP15, USE1, WSB2, GPRC5C, CENPE, THSD1, ACSS2, NXF2, MYDGF, PDGFC, PCDHGA7, PCDHG@, PCDHA@, PCDHB3, H2AJ, CETP, RNF20, ARFGAP1, LGR4, HES6, MACROH2A2, GPRC5D, KLK15, ENOSF1, CDV3, UBE2Q1, TENT5A, PRR5, THUMPD1, ZNF407, CHD7, HIF1AN, KDM4D, CEP72, CFL1, BTBD7, VPS35, GSDMC, PARD3, CNC2, ADAMTS9, CIAPIN1, TWSG1, KNL1, RAB25, APMAP, SCYL3, SLURP1, SMURF1, ZNFX1, VANGL2, LYRM2, SMAGP, ZNF304, GJD2, BIRC6, TENM2, PPM1H, CRACD, ARID1B, CDKN2D, CTNNBIP1, LTB4R2, CDS1, CTPS2, CEBPD, CHST7, DIABLO, KCNK12, AKIP1, JPH1, FMN2, CEBPA, TCIM, DPYSL5, NDUFA4L2, C1GALT1, EXOSC5, PMEPA1, DUSP22, EMSY, POGLUT1, CDC42SE2, TMEM176A, CHDH, CHD4, UGT2B28, RBM47, PCDH18, EXOSC4, TPP1, ROBO4, DLL4, CLIC1, EGLN1, UGT1A9, TMEM106B, CRLS1, CLC, RBFOX1, MPHOSPH8, EPDR1, FBLIM1, IL17RD, TRIM44, PPP1R12C, SMOX, CHPF2, VPS53, RNF216, ARID4B, LUC7L3, LTB4R, KDM3B, GALNT7, BCL11A, IL17D, MYO3A, SLC37A1, CCR4, CCR3, DCUN1D1, ERRFI1, TREM2, SOX18, CYTL1, WNT4, SSH1, MIEF1, RNF111, TRPM4, DCHS2, AHI1, CHRNA7, TMEM45A, FEZF2, KIF26B, AKIRIN2, PIWIL2, TRIM68, MTPAP, CEP55, RIF1, PBRM1, APPL2, CHIT1, MAP1S, DPPA4, SLC38A7, STEAP3, PACC1, IMP3, ANKRD40CL, CHUK, GID8, SLC66A1, NDE1, ERCC6L, CKMT1B, CDHR2, NXPE4, SYTL2, NSUN2, ALKBH5, CASZ1, CT55, CDKAL1, NSD3, FOCAD, TRMT10C, LPCAT2, CKB, SSH3, TIPIN, NTSR2, DAPK2, CYBA, TANK, TROAP, PIGK, DNAJB6, SMC4, EFNB1, DNM1L, NR1H3, GPC6, EEF2, ARPC4, ACTR2, KIF20A, HIPK3, LRPPRC, EDN3, ABI2, S1PR1, SLC25A13, TNK2, BCL2L10, CELSR3, TSHZ1, PTBP3, TRIM66, DDX46, KBTBD11, MFN2, MEGF6, DCLRE1A, WDR1, HS3ST2, USP3, SLC23A2, FGF19, TNFSF15, MED12, MED13, NUP153, CCS, FGFBP1, THOC1, RCE1, TXNDC9, PSME3, HYOU1, DUSP8, NDC80, WFDC2, IFITM3, OLFM1, ACAA2, HAX1, MERTK, SLC30A9, DSC1, TIMM44, SEC23A, CAP1, STK25, NCOA2, SEMA3C, FBLN5, CIB2, ARID3A, DDX17, TUBB4B, DUSP9, NME6, DVL1, ECM1, EIF1, FLOT1, CD96, NBR2, ECE1, RABEPK, SPRY1, EBF1, IRX5, RAMP3, ZMPSTE24, STUB1, STAG1, LILRB2, DLEU1, VPS26C, LANCL1, DYRK1A, MAGI2, FARP2, MFAP3L, SLC9A3R2, ENPEP, KIF3B, TM9SF2, SELENOF, TJP2, CYP7B1, MYLKP1, ELK4, ELK3, HOMER2, ELK1, HAND2, ELF3, ONECUT2, KIF23, TBX4, PAGE4, ADAMTS1, ELANE, TXNL1, RPL23, ZBTB24, DMTN, FIBP, LRRFIP1, MSC, RPS6KA5, CACNA2D2, AIMP1, MFHAS1, STK17B, STK17A, CYTH2, COPB2, GPR55, EPHA4, STOM, TRIP13, TRIP10, B4GALT6, B4GALT5, TCEAL1, GOSR1, BAG4, GMFG, EI24, MICAL2, ZNF516, VGLL4, BZW1, ULK2, EIF4A2, USP6NL, SEMA3E, EIF4A1, EIF2S3, SUSD6, PCLAF, EIF2S1, BMS1, TOMM20, SCRN1, TSC22D2, RB1CC1, EGR2, SH3PXD2A, IKBKE, CLCA2, KIF20B, TP53I3, IGDCC3, CHD1L, CCL4L2, BCAR1, GTF2IRD1, SOX13, RBM39, CREB5, SEC24C, PRDX6, EIF5, PITPNM1, RIN1, NCOR1, RNF7, EIF4EBP2, KLK4, DEAF1, IPO7, CASP14, CARD8, SEPHS1, CCT5, DDX1, GADD45A, AKR1C1, RAB21, PALLD, DAXX, PDS5B, NMNAT2, ZNF609, GGA2, SETD1B, MYCBP2, PHF8, KDM6B, EPB41L3, LRCH1, CIC, DDX6, RPH3A, SEPTIN6, DIS3, PDAP1, TUSC2, CBX3, U2AF2, EXOSC8, DEFB1, LAMB4, MRAS, DEFA6, RAB11FIP2, DEFA1, CPEB3, CHSY1, ADGRL1, NLRP1, ATG14, DDX11, MORC2, DHX9, DGKG, IFNGR2, NOP56, NOMO1, SF3B1, HEY1, TRAM1, ETHE1, SEC11A, PES1, CBX7, SPIDR, SRRM2, CYP3A7, SEC14L2, MAPK8IP2, DNPEP, PSD4, RASD2, TSPAN12, PIGN, GTPBP4, PADI4, RHOQ, ADNP, MESD, MAU2, RCOR1, DAB2, ARL6IP1, RRS1, SULF1, NUP210, DAB1, ARC, PACS2, BOP1, ATP11A, ANKRD12, CAMTA1, CYP51A1, ICOSLG, TRIM2, TTC28, LARP1, PPP1R13B, PHB2, COPE, PARK7, STAG2, RAI2, MASP2, TRDMT1, GIPC1, NES, KDM5B, SRSF10, IQGAP2, VAMP5, CCR9, DNM2, WASF3, UTP14A, DYNC1H1, PDE10A, CYP46A1, NMU, MRPS30, MALT1, SIX2, OGA, SLC6A14, FUT9, BATF, GLRX3, ARL6IP5, SLC19A2, CCT7, CCT2, TXNRD2, SMC2, ERN2, DPH1, SLCO1B1, PAICS, RBCK1, IVNS1ABP, DOK1, IGF2BP3, SPINT2, TNFSF13B, EBP, MTMR11, PAPOLA, MAGED2, FASTK, CD160, DIAPH1, KAT7, PLAAT3, HHLA2, DHODH, NUDT6, NUDT5, DGUOK, LZTS1, MAP4K1, CELF3, PTENP1, CFD, KLK8, TREX2, GALNT5, RASSF8, PDCD10, DIO2, BTN3A1, FGFR1OP, ATF7, ERP29, MLLT11, STIP1, EBNA1BP2, FERMT2, METAP2, SLC27A4, LILRB4, TENT4A, PRDM5, ADRM1, WWP1, DEPP1, TRIOBP, DIDO1, WDR5, ADAMTS13, ADAMTS8, H3P40
-
Telomere Length, Mean Leukocyte
Omim
Heritability of age-dependent attrition rate was estimated at 28%. Individual unique environmental factors, estimated at 72%, affected attrition rate.
-
Epileptic Encephalopathy, Early Infantile, 4
Omim
Using array CGH, Saitsu et al. (2012) identified 2 different de novo heterozygous deletions involving the STXBP1 gene in 2 (7.1%) of 28 patients with cryptogenic early-onset epileptic encephalopathy.SCN1A, SCN9A, STXBP1, SCN2A, SCN1B, PCDH19, GABRG2, GABRA1, TNRC6A, POMC, PMP22, RAPGEF2, SAMD12, KCNQ2, SCN8A, ADRA1D, PVALB, POLG, SCN2B, KCNQ3, SST, GABRB3, GPR55, PRRT2, TLR1, VIP, LINC01672, CPLX1, EPM2A, B3GNT2, OPN1MW2, SLC12A5, LOH19CR1, GPHN, ARX, EJM2, B3GNTL1, CACNA1G, TSPYL4, ACHE, CDKL5, FOXM1, ALDH7A1, CACNA1A, CACNB4, CHD2, CSTB, CYP2C19, E2F1, EPHA5, FOXG1, MTOR, SLC12A2, GABRA2, GABRD, GAPDH, OPN1MW, IGF1, MECP2, RPS19, SCN7A, SLC2A1, OPN1MW3
-
Inflammatory Bowel Disease 3
Omim
Hampe et al. (1999) reported on a 2-stage linkage and association analysis of both a basic population of 353 affected sib pairs and an extension of this population to 428 white affected sib pairs of northern European extraction. Genotyping 28 microsatellite markers on chromosome 6, they found a peak multipoint lod score of 4.2 at D6S461 for the IBD phenotype.
-
Psoriasis 4, Susceptibility To
Omim
Results consistent with these data were obtained by typing an independent sample that included 28 patients and 56 controls originating from Sardinia.