-
Lethal White Syndrome
Wikipedia
[ edit ] See also: Autosomal recessive § Nomenclature of recessiveness Lethal white syndrome has been described by researchers as both dominantly and recessively inherited. [27] [28] [29] Lethal white syndrome is described as recessive because heterozygotes (written Oo or N/O ) are not affected by intestinal agangliosis.
- Ephebiphobia Wikipedia
-
Amyloidosis
Wikipedia
Chemotherapy and steroids, with melphalan plus dexamethasone , is mainstay treatment in AL people not eligible for transplant. [19] In AA, symptoms may improve if the underlying condition is treated; eprodisate has been shown to slow renal impairment by inhibiting polymerization of amyloid fibrils. [ medical citation needed ] Management of ATTR amyloidosis will depend on its classification as wild type or variant. [27] Both may be treated with tafamidis , a low toxicity oral agent that prevents destabilization of correctly folded protein. [27] Studies showed tafamidis reduced mortality and hospitalization due to heart failure . [27] Previously, for variant ATTR amyloidosis, liver transplant was the only effective treatment. [27] However, newer therapies including diflunisal , an anti-inflammatory drug, and inotersen and patisiran , drugs which prevent misfolded protein formation, have shown early promises in slowing disease progression. [27] The latter two drugs have shown their benefit in neurological impairment scores and quality of life measures. [27] However, their role in cardiac ATTR amyloidosis is still being investigated. [27] In 2018, patisiran was not recommended by NICE in the UK for hereditary transthyretin-related amyloidosis. [28] As of July 2019 further review however is occurring. [29] It was approved for this use in the United States however. [30] Support groups [ edit ] People affected by amyloidosis are supported by organizations, including the Amyloidosis Research Consortium, Amyloidosis Foundation, Amyloidosis Support Groups, and Amyloidosis Australia. [31] [32] Prognosis [ edit ] Prognosis varies with the type of amyloidosis and the affected organ system.TTR, PSEN1, GSN, APP, ACHE, APOE, HMOX1, APOC3, ZDHHC13, APOC2, TNFRSF1A, CCND1, POLA1, B2M, BCHE, BDNF, NLRP3, PRNP, OSMR, CASP3, MME, CSF2, CST3, SAA2, MEFV, MAPT, LYZ, LIG4, LAMC2, IL6, IL1B, TREM2, IDE, BACE1, IAPP, SAA1, GFAP, APCS, TNF, AGER, SNCA, APOA1, NEFL, TGFBI, CLU, ADAM10, IL1A, SUCLA2, DPYD, IL4, NGF, NFE2L2, APOA2, LPAL2, ABCB1, BECN1, PPARG, NRGN, TARDBP, BIN1, BCL2, TLR4, MMP9, ABCA7, CTSD, DCLRE1B, LRP1, TYROBP, SIRT1, TGFB1, LECT2, CHI3L1, MOK, RELN, PSEN2, S100A9, TSPO, LEP, CTSB, SOD1, SYP, ALOX5, KHDRBS1, DLG4, MCIDAS, CAT, CRP, IGF1, FGA, SMUG1, NUP62, DCTN4, HP, S100B, GTF2H1, GSK3B, SOD2, AQP4, CHRNA4, VEGFA, PIK3CD, ABCB6, CDK5, PLG, SQSTM1, PIK3CG, PIK3CB, MAPK3, ITM2B, PIK3CA, SORL1, IL10, LPA, VSNL1, ROS1, RXRA, PTPRC, EGR1, PARP1, ALB, CD40, STS, TLR2, PLD3, CD36, GABPA, HTT, SDC1, PYCARD, SH2D1A, LINC01672, ACE, MAPK8, CNR2, MAP3K5, CYP46A1, TSHZ1, CD55, PINK1, SERPINA3, ADIPOQ, GRM5, HTRA1, MAPK1, TRPC6, DYSF, TYRP1, RNR2, UBQLN1, QPCT, NCAM1, NFIB, MMP2, HLA-DRB1, NFIA, HCRT, PPARA, HDAC3, UCHL1, ABCG1, MAP2, PTGS2, PPARGC1A, PDB1, LDLR, NFIC, LBP, TXN, DKK1, INSR, SYNM, NCSTN, IL17A, IL13, GH1, P2RX7, RBP4, IL1RN, MFAP1, PDE5A, NFIX, A2M, CD40LG, FYN, NGB, DDIT3, CRYAB, CD38, APLP2, C4BPA, MIR200A, ARG1, ESCO1, EPO, DDR1, CYLD, CALCA, FCGRT, FAP, DECR1, LRRK2, APRT, CR1, GCG, DYRK1A, CHRM3, GDNF, ECE1, EDN1, ACTB, DRD1, IGAN1, TNMD, CHAT, DPYSL2, BAX, MFT2, PCSK9, GCSAM, LRRTM3, STOX1, CDK5R1, SUMO4, GOLGA6A, OSTN, ANO5, LOC390714, ARHGEF7, DUOX2, CD163, UBR1, FSIP1, SV2A, SNAP91, MAGI2, ZEB2, KEAP1, UCN3, TOMM20, PCLAF, CLSTN3, HDAC9, CD109, ACTRT1, CARTPT, MAD2L1BP, BCAR1, GDF15, TRIM69, TMC4, GRAP2, TICAM1, RMDN2, DAB2IP, CRADD, MSC, SLC2A12, RAB11A, MIR137, MIR107, RIPK1, VWF, DEFB103A, VTN, DNM1P33, LOC643387, SCFV, VIP, SFTPA1, VDR, VCAM1, UTRN, NR1H2, C20orf181, BACE1-AS, OCLN, TYRO3, MICA, TMX2-CTNND1, LOC102723407, LOC102724971, LINC02210-CRHR1, UPK3B, HSP90B2P, TRAF6, LOC105379528, WNT1, XBP1, XK, AGPS, MIR132, URI1, WDR1, EIF3A, MIR15B, MIR181C, UNC5C, DENR, DEGS1, MIR29A, MIR29B1, CST7, LRP8, ULK1, AXIN1, PICALM, MIR29C, BAS, FXR1, PSCA, AIMP2, TFEB, PLA2G7, NPHS2, DBA2, PPIF, DGCR2, RAB21, TP53, HSPA14, DCDC2, GDE1, DUOX1, GSAP, DAPK2, TLR9, TREM1, QPCTL, DDAH1, ODAM, SLC25A38, RMDN3, RCBTB1, SYBU, SIRT3, USE1, DEFB103B, CRTC1, CTNNBL1, ARC, SLC17A7, ARL6IP1, TBC1D9, PRDX5, BACE2, RNF19A, ADIPOR1, SNX12, EEF2K, SNX8, FLVCR1, TMEM176B, F11R, IGKV2-29, IGKV3-20, ASCC1, PRLH, GAL, IGKV1D-8, SH2B1, IGKV3D-15, IGHV3-69-1, IGHV3OR16-7, APH1A, HTRA2, VPS4A, RMDN1, HSPB8, POLDIP2, SDF4, CHMP2B, KIF1B, SHANK2, HDAC5, ATF6, GPNMB, OLFM1, PRMT5, PHF6, RTN3, COL25A1, LILRB2, CALCOCO2, MFSD2A, ABI2, NAV3, CHRFAM7A, BMF, NLRP12, BMS1P20, OPTN, PRRT2, MBL3P, NR1H3, DNM1L, CDCA5, SH2D3C, TOM1, BCL2L11, PDCD6IP, CIB1, MAP1LC3B, KAT5, FERMT2, NLGN1, NLRP1, HPS5, CENPK, KLK8, WDHD1, TPPP, ABCG4, RIPK3, HHIP, SDS, STIP1, SLC8B1, CLEC7A, UBE2Z, EDAR, CARD14, MMEL1, CLDN16, PAGR1, TXNIP, EHMT1, AHSA1, ATG7, TPSG1, PPP3R1, CLDN5, EREG, NR5A1, MTOR, FPR1, FN1, FOXO1, FGF2, FCN2, EFEMP1, FAT1, FANCD2, F10, F3, ESR2, ESR1, EPHB2, TLR5, EPHA3, EPHA1, ELN, SERPINB1, EHHADH, E2F1, DUSP6, DPP4, DPEP1, DNMT3A, DNMT1, DNM1, DLX4, DLG2, GAST, FUS, G6PD, GALNS, HSPA9, HSPA4, HSF1, HSD11B1, FOXA2, NR4A1, HMGB1, HMBS, HLA-DMA, NRG1, HFE, HDAC2, GSK3A, CXCL2, GRM3, GRM2, GRIN2B, GRIN1, GRN, GRIA1, GPX1, GNA12, GEM, GDF2, GCHFR, GC, GATA6, GAS6, GAPDH, DES, DCX, DCN, C1QB, C1QBP, BSG, BRCA2, BGN, BCR, TNFRSF17, AVP, ATF4, SERPINC1, ARSA, ABCC6, FAS, APOA4, APLP1, BIRC3, APBB2, APAF1, ANXA5, ANXA1, ALOX15, ALOX12, AKT1, AIF1, AGTR1, AGT, ACAN, ACAT1, ABCA4, ABCA1, C1QA, C1QC, CYBB, C3, CTNND1, CSF3, CSE1L, MAPK14, CRMP1, CRK, CRHR1, CRH, CPB2, COL11A2, CLCN3, CHIT1, AKR1C4, CETN1, CEBPA, CDR1, CD74, CD68, CD33, CD19, CD14, RUNX1T1, CAV1, CASR, CASP1, CAMK2G, CAMK4, CACNA1C, C4BPB, HSP90AA1, HSPD1, HSPG2, MAPK12, S100A8, S100A6, SORT1, S100A1, RPL29, RHD, RET, RARB, PVALB, PTX3, PTGS1, MAP2K6, MAP2K1, PRKCB, PRKCA, PPY, PTPA, PPID, PPARD, POR, POLB, PMP22, PLK1, PLA2G4A, PITX3, PIN1, SERPINB9, SERPINB6, SLC25A3, SAA4, ATXN7, PAWR, SCD, TIMP1, TGFBR2, PRDX2, TAT, SYT1, SYN1, VAMP1, STK11, ST13, SST, TRIM21, SPRR2A, SPP1, SOAT1, SNCG, SMN2, SMN1, SLC6A3, SLC5A5, SLC1A3, PMEL, SH3GL2, SGCG, SRSF2, SEMG1, SELPLG, CXCL12, CCL5, SCT, SERPINF1, REG3A, HTR1B, MAZ, MAOA, MAG, TACSTD2, LTB, LNPEP, LMNB1, LIMK1, LCN2, LAIR1, KRAS, KLC1, KNG1, KIR3DL1, ITIH4, ITGAM, IRS1, PDX1, INS, CXCL10, ING1, IL18, IL12A, CXCR2, CXCL8, IL2, IFNG, IFNB1, ID2, HTR2A, MATN1, MBL2, SERPINE1, MBP, PAEP, P2RY2, P2RX4, OCA2, NTRK2, YBX1, SLC11A2, NPTX1, NPPA, NPC1, NPY, NOTCH1, NOS3, NFKB1, NEFH, NCF2, NAGLU, MYOC, COX1, MT3, ABCC1, MPZ, MPV17, MNAT1, MMP3, AFF1, MFGE8, DNAJB9, CHST6, H3P40
-
Evil Eye
Wikipedia
Reciting Sura Ikhlas , Sura Al-Falaq and Sura Al-Nas from the Qur'an , three times after Fajr and after Maghrib is also used as a means of personal protection against the evil eye. [24] [ better source needed ] [ failed verification ] Salafi scholars have pointed some conditions from the Quran and Hadith , which includes performing exorcism using the words of God or his names, reciting in Arabic or in a language which can be understood by the people, not using any talismans or amulets or fortune-tellers or any magic, nor asking jinns to help. [25] [26] [27] [28] Scholars have difference of opinion whether talismans using the Quran is permissible or not. [25] Assyrians [ edit ] A Ruby Eye Pendant from an ancient civilization in Mesopotamia was possibly used as an amulet to protect against the evil eyes.
-
Spinocerebellar Ataxia 6
Omim
In their series of 32 patients, onset was usually late and the (CAG)n stretch varied between 22 and 28 trinucleotide units, the shortest trinucleotide repeat expansion causing spinocerebellar ataxia.CACNA1A, ATXN2, ATXN1, ATXN7, PPP2R2B, WWOX, TRPC3, CACNA1G, RUBCN, STUB1, NOP56, AFG3L2, CIC, TDP2, SPTBN2, ANO10, SNX14, SCYL1, ELOVL5, UBA5, RBM17, TTBK2, VWA3B, SYT14, ATXN1L, TGM6, CWF19L1, CCDC88C, GRM1, PRKCG, FOXC1, GFI1, MME, GRID2, ATXN3, PDYN, LY6E, CASP3, TK2, BEAN1, MPI, HSF1, GTF2H1, PVALB, HSPA1A, DAP, CALB1, CACNA1F, TWNK, DCTN4, HSPA1B, DNAJB1, IL1B, ATXN8OS, NUP62, KHDRBS1, MYD88, SQSTM1, TPO, TNF, BDNF, SLC6A3, SLC2A1, AR
-
Familial Hypercholesterolemia
Wikipedia
In the more serious homozygous forms, the receptor is not expressed at all. [3] Some studies of FH cohorts suggest that additional risk factors are generally at play when a person develops atherosclerosis. [17] [18] In addition to the classic risk factors such as smoking, high blood pressure, and diabetes, genetic studies have shown that a common abnormality in the prothrombin gene (G20210A) increases the risk of cardiovascular events in people with FH. [19] Several studies found that a high level of lipoprotein(a) was an additional risk factor for ischemic heart disease. [20] [21] The risk was also found to be higher in people with a specific genotype of the angiotensin-converting enzyme (ACE). [22] Screening [ edit ] Screening among family members of people with known FH is cost-effective . [23] Other strategies such as universal screening at the age of 16 were suggested in 2001. [24] [25] The latter approach may however be less cost-effective in the short term. [26] Screening at an age lower than 16 was thought likely to lead to an unacceptably high rate of false positives . [5] A 2007 meta-analysis found that "the proposed strategy of screening children and parents for familial hypercholesterolaemia could have considerable impact in preventing the medical consequences of this disorder in two generations simultaneously." [27] "The use of total cholesterol alone may best discriminate between people with and without FH between the ages of 1 to 9 years." [28] [27] Screening of toddlers has been suggested, and results of a trial on 10,000 one-year-olds were published in 2016. ... "Medicine 1985" . Retrieved 2008-02-28 . External links [ edit ] MedicinePlus: Familial Hypercholesterolemia Classification D ICD - 10 : E78.0 ICD - 10-CM : E78.01 ICD - 9-CM : 272.0 OMIM : 143890 MeSH : D006938 DiseasesDB : 4707 External resources MedlinePlus : 000392 eMedicine : med/1072 v t e Inborn error of lipid metabolism : dyslipidemia Hyperlipidemia Hypercholesterolemia / Hypertriglyceridemia Lipoprotein lipase deficiency/Type Ia Familial apoprotein CII deficiency/Type Ib Familial hypercholesterolemia/Type IIa Combined hyperlipidemia/Type IIb Familial dysbetalipoproteinemia/Type III Familial hypertriglyceridemia/Type IV Xanthoma/Xanthomatosis Hypolipoproteinemia Hypoalphalipoproteinemia/HDL Lecithin cholesterol acyltransferase deficiency Tangier disease Hypobetalipoproteinemia/LDL Abetalipoproteinemia Apolipoprotein B deficiency Chylomicron retention disease Lipodystrophy Barraquer–Simons syndrome Other Lipomatosis Adiposis dolorosa Lipoid proteinosis APOA1 familial renal amyloidosis v t e Cell surface receptor deficiencies G protein-coupled receptor (including hormone ) Class A TSHR ( Congenital hypothyroidism 1 ) LHCGR ( Luteinizing hormone insensitivity , Leydig cell hypoplasia , Male-limited precocious puberty ) FSHR ( Follicle-stimulating hormone insensitivity , XX gonadal dysgenesis ) GnRHR ( Gonadotropin-releasing hormone insensitivity ) EDNRB ( ABCD syndrome , Waardenburg syndrome 4a , Hirschsprung's disease 2 ) AVPR2 ( Nephrogenic diabetes insipidus 1 ) PTGER2 ( Aspirin-induced asthma ) Class B PTH1R ( Jansen's metaphyseal chondrodysplasia ) Class C CASR ( Familial hypocalciuric hypercalcemia ) Class F FZD4 ( Familial exudative vitreoretinopathy 1 ) Enzyme-linked receptor (including growth factor ) RTK ROR2 ( Robinow syndrome ) FGFR1 ( Pfeiffer syndrome , KAL2 Kallmann syndrome ) FGFR2 ( Apert syndrome , Antley–Bixler syndrome , Pfeiffer syndrome , Crouzon syndrome , Jackson–Weiss syndrome ) FGFR3 ( Achondroplasia , Hypochondroplasia , Thanatophoric dysplasia , Muenke syndrome ) INSR ( Donohue syndrome Rabson–Mendenhall syndrome ) NTRK1 ( Congenital insensitivity to pain with anhidrosis ) KIT ( KIT Piebaldism , Gastrointestinal stromal tumor ) STPK AMHR2 ( Persistent Müllerian duct syndrome II ) TGF beta receptors : Endoglin / Alk-1 / SMAD4 ( Hereditary hemorrhagic telangiectasia ) TGFBR1 / TGFBR2 ( Loeys–Dietz syndrome ) GC GUCY2D ( Leber's congenital amaurosis 1 ) JAK-STAT Type I cytokine receptor : GH ( Laron syndrome ) CSF2RA ( Surfactant metabolism dysfunction 4 ) MPL ( Congenital amegakaryocytic thrombocytopenia ) TNF receptor TNFRSF1A ( TNF receptor associated periodic syndrome ) TNFRSF13B ( Selective immunoglobulin A deficiency 2 ) TNFRSF5 ( Hyper-IgM syndrome type 3 ) TNFRSF13C ( CVID4 ) TNFRSF13B ( CVID2 ) TNFRSF6 ( Autoimmune lymphoproliferative syndrome 1A ) Lipid receptor LRP : LRP2 ( Donnai–Barrow syndrome ) LRP4 ( Cenani–Lenz syndactylism ) LRP5 ( Worth syndrome , Familial exudative vitreoretinopathy 4 , Osteopetrosis 1 ) LDLR ( LDLR Familial hypercholesterolemia ) Other/ungrouped Immunoglobulin superfamily : AGM3, 6 Integrin : LAD1 Glanzmann's thrombasthenia Junctional epidermolysis bullosa with pyloric atresia EDAR ( EDAR hypohidrotic ectodermal dysplasia ) PTCH1 ( Nevoid basal-cell carcinoma syndrome ) BMPR1A ( BMPR1A juvenile polyposis syndrome ) IL2RG ( X-linked severe combined immunodeficiency ) See also cell surface receptorsLDLR, APOB, PCSK9, LDLRAP1, APOE, CETP, LPL, PON1, HMGCR, ABCG5, ABCA1, APOA4, PON2, ABCG8, LIPC, SREBF2, APOA2, APOC3, GK, STAP1, ZNF823, PDE4A, TYK2, ZNF846, PRKCSH, ZNF561, FARSA, ZNF561-AS1, ICAM3, MIR6886, RAVER1, ZNF317, C3P1, KANK2, LPA, CACNA1A, ZNF562, CARM1, CDC37, ZNF563, APOA1, EGF, CRP, SLCO1B1, CYP7A1, TNF, ANGPTL3, CAD, LIPA, CHDH, COL9A2, COL9A3, COMP, ABCB1, SELE, SCN8A, COL9A1, LRP1, LCAT, REN, MMP9, MYBPC3, NHS, OCA2, SERPINE1, CCN6, SLC2A1, PEA15, THBD, PLA2G7, VLDLR, AGT, CXCR1, MIR505, AGO2, CXCL8, ACE, VSX1, F3, CCR4, HLP, HPD, RUNX1, NR3C1, MIR33A, ALOX5AP, FTH1, ADPRH, SACS, TNFRSF10C, TNFRSF10A, TNFSF10, PHACTR1, TNFRSF10D, KHSRP, LINC01194, PDE5A, APOA5, APOL1, HMGA2, FGF23, LINC01672, USF2, USF1, PNPLA5, ADIPOQ, CH25H, TNFRSF13C, MYLIP, INSIG2, HDL3, PLA2G15, TBC1D9, COG2, PSIP1, TNFSF13B, ACSS2, PLAAT1, AICDA, KIDINS220, TRIB1, MVP, SEMA6A, ABCG2, LIPG, ADRB3, ACCS, SIGLEC7, CD163, TYRP1, ROS1, TXN, TNFSF4, GH1, GHR, GLB1, GCLC, GPT, GPX4, CXCL3, GSS, HCLS1, CFH, HSPG2, ICAM1, IL1B, IL2RB, IL4R, FGB, EXT2, EXT1, ANXA5, APOC2, APOH, BTF3P11, C4BPA, C4BPB, CD28, COX7B, ESR1, CRABP2, CUX1, EDN1, CELSR2, EGFR, EPHX2, IL6, IL10, IL18, SP1, S100A12, SCNN1A, SDC2, SLC22A3, SMARCA4, SORL1, STAT1, POR, TCF3, TGFB1, TIMP1, TLR4, TNFRSF1B, TNNT1, PTPRC, ABO, ITGB2, MMP1, KDR, STMN1, ALOX5, ABCD2, LRP5, KMT2A, MMP2, TNFRSF11B, MMP3, MPO, MTHFR, NOS3, NPPA, OLR1, DALIR
-
Swine Influenza
Wikipedia
Additional tests of 150 pigs outside the area were negative. [28] [29] Structure [ edit ] Structure of H1N1 Virion. ... "An overview of swine influenza" . Vet Q . 28 (2): 46–53. doi : 10.1080/01652176.2006.9695207 . ... PMID 17366454 . ^ "Symptoms of H1N1 (Swine Flu)" . YouTube. 2009-04-28 . Retrieved 2011-05-22 . ^ a b Centers for Disease Control and Prevention (April 27, 2009). ... PMID 15380362 . ^ World Health Organization (28 October 2005). "H5N1 avian influenza: timeline" (PDF) . ... "Influenza: evolving strategies in treatment and prevention". Semin Respir Crit Care Med . 28 (2): 144–58. doi : 10.1055/s-2007-976487 .ABCF1, APLP1, TEKT1, L3MBTL3, BUD13, MEDAG, CFAP74, RRP7BP, ARAP2, PNLIPRP3, ENPP6, PXDNL, SLITRK4, CEP128, LINGO2, SPRED2, SERHL2, TMPRSS11F, TEX26-AS1, LINC01811, LINC02334, CEP290, ESYT2, MAN1C1, NUMA1, BRAF, DLG2, DMD, ACSL3, HLA-DQA2, AGFG1, MITF, MMP9, PIP4K2A, TBC1D2, PTPRR, ADAM12, GTF2IRD1, TUBGCP2, SPINK4, LRP1B, ATAD2B, GIPC2, LINC02336
-
Waardenburg Syndrome
Wikipedia
It leads to the conclusion that the double mutation of MITF is associated with the extremity of Waardenburg syndrome and may affect the phenotypes or symptoms of the syndrome. [28] Classification table [ edit ] Type OMIM Gene Locus Inheritance Type 1 (WS1) 193500 PAX3 2q36.1 [29] Autosomal dominant Type 2A (WS2A, originally WS2) 193510 MITF 3p14.1–p12.3 Autosomal dominant Type 2B (WS2B) 600193 WS2B 1p21–p13.3 Autosomal dominant Type 2C (WS2C) 606662 WS2C 8p23 Autosomal dominant Type 2D (WS2D) 608890 SNAI2 8q11 Autosomal recessive Type 2E (WS2E) 611584 SOX10 22q13.1 Autosomal dominant Type 3 (WS3) 148820 PAX3 2q36.1 Autosomal dominant or autosomal recessive Type 4A (WS4A) 277580 EDNRB 13q22 Autosomal dominant or autosomal recessive Type 4B (WS4B) 613265 EDN3 20q13 Autosomal dominant or autosomal recessive Type 4C (WS4C) 613266 SOX10 22q13.1 Autosomal dominant Treatment [ edit ] There is currently no treatment or cure for Waardenburg syndrome. ... ' " . BBC News . Retrieved 2018-01-28 . ^ MARKAKIS, MARIOS N.; SOEDRING, VIBEKE E.; DANTZER, VIBEKE; CHRISTENSEN, KNUD; ANISTOROAEI, RAZVAN (2014-08-01).
-
Anaphylaxis
Wikipedia
The most common are β-lactam antibiotics (such as penicillin ) followed by aspirin and NSAIDs . [13] [25] Other antibiotics are implicated less frequently. [25] Anaphylactic reactions to NSAIDs are either agent specific or occur among those that are structurally similar meaning that those who are allergic to one NSAID can typically tolerate a different one or different group of NSAIDs. [26] Other relatively common causes include chemotherapy , vaccines , protamine and herbal preparations. [3] Some medications ( vancomycin , morphine , x-ray contrast among others) cause anaphylaxis by directly triggering mast cell degranulation. [8] The frequency of a reaction to an agent partly depends on the frequency of its use and partly on its intrinsic properties. [27] Anaphylaxis to penicillin or cephalosporins occurs only after it binds to proteins inside the body with some agents binding more easily than others. [12] Anaphylaxis to penicillin occurs once in every 2,000 to 10,000 courses of treatment, with death occurring in fewer than one in every 50,000 courses of treatment. [12] Anaphylaxis to aspirin and NSAIDs occurs in about one in every 50,000 persons. [12] If someone has a reaction to penicillin, his or her risk of a reaction to cephalosporins is greater but still less than one in 1,000. [12] The old radiocontrast agents caused reactions in 1% of cases, while the newer lower osmolar agents cause reactions in 0.04% of cases. [27] Venom [ edit ] Venom from stinging or biting insects such as Hymenoptera (ants, bees, and wasps) or Triatominae (kissing bugs) may cause anaphylaxis in susceptible people. [7] [28] [29] Previous reactions, that are anything more than a local reaction around the site of the sting, are a risk factor for future anaphylaxis; [30] [31] however, half of fatalities have had no previous systemic reaction. [32] Risk factors [ edit ] People with atopic diseases such as asthma , eczema , or allergic rhinitis are at high risk of anaphylaxis from food, latex , and radiocontrast agents but not from injectable medications or stings. [3] [8] One study in children found that 60% had a history of previous atopic diseases, and of children who die from anaphylaxis, more than 90% have asthma. [8] Those with mastocytosis or of a higher socioeconomic status are at increased risk. [3] [8] The longer the time since the last exposure to the agent in question, the lower the risk. [12] Pathophysiology [ edit ] Anaphylaxis is a severe allergic reaction of rapid onset affecting many body systems . [5] [6] It is due to the release of inflammatory mediators and cytokines from mast cells and basophils , typically due to an immunologic reaction but sometimes non-immunologic mechanism. [6] Interleukin (IL)–4 and IL-13 are cytokines important in the initial generation of antibody and inflammatory cell responses to anaphylaxis.KNG1, ASPG, LTB4R, NLRP3, BTK, PTGS1, PTGS2, ADRB1, VCAM1, TPSD1, ACE, F9, VWF, CD63, FCGR2B, HP, MRGPRX2, FCGR3A, FCGR2A, STK11, MILR1, CCL2, IL10, IL33, TNF, FCGR1A, OGA, FCER1A, IL6, CD33, SIRT1, IL9, FCGR3B, IL4, MCAT, IL5, MCPH1, STAT3, IGHE, SLC16A1, ADRB2, COX8A, PTPN1, VEGFA, RHD, TRPV1, ABHD16A, NR0B2, TRPC1, SCN5A, SAA1, PVALB, RPE65, TM7SF2, STXBP2, RGS13, STAT6, BHLHE40, SPG7, CCL7, SAA2, CCL3, PTPN6, ABL1, SOCS1, NT5C3A, TNFRSF12A, ARHGEF40, WDR11, CYSLTR2, AICDA, AHRR, CABS1, TSLP, SESTD1, C20orf181, LINC01672, LOC102723407, LOC102723971, LOC102724971, LINC02210-CRHR1, BFAR, CPA4, TNFSF10, IGHV3-69-1, HDAC3, SOCS3, HACD1, PCLAF, NR1H4, IL18BP, CAP1, PROC, SORBS1, MASP2, BRD4, PART1, CHIC2, SGSM3, IGHV3OR16-7, PTAFR, MTTP, PLSCR1, RCAN1, CPN1, CRH, CRHR1, CSF3, CTAA1, CTSD, DNASE1, ELAVL2, CHRM3, EPO, ESR1, ESR2, F8, F10, MS4A2, FCGR1B, CPA3, CDH15, PITX2, APOA1, ACADM, ADAM10, JAG1, AGT, AGXT, AHR, ANGPT1, FASLG, CD36, ARSA, AVP, BAAT, BRAF, TSPO, CD80, CD86, FLG, GAPDH, GATA1, NFKB1, LNPEP, LTC4S, LYZ, MITF, MPO, MRC1, ABO, NHS, GATA2, NOS3, NOTCH1, PAEP, PAFAH1B2, PECAM1, PFAS, SERPINB6, KIT, ISG20, IL18, IL17A, GCG, GPT, HDC, HLA-DQB1, NR4A1, HSPA4, HTR3A, ICAM1, IFNG, IGH, IL1A, IL1B, IL2RA, IL4R, IL13, PERCC1
-
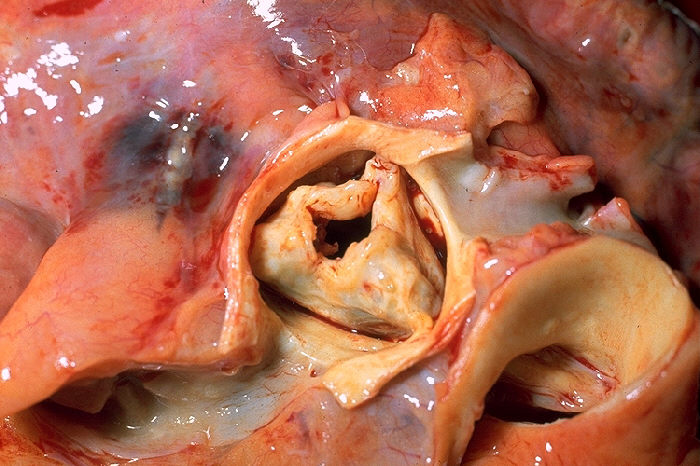
Aortic Stenosis
Wikipedia
It can also become necessary for the treatment of an aortic aneurysm, less frequently for congenital aortic stenosis. [28] Aortic valve replacement [ edit ] Main article: aortic valve replacement In adults, symptomatic severe aortic stenosis usually requires aortic valve replacement (AVR). [3] While AVR has been the standard of care for aortic stenosis for several decades, As of 2016 [update] aortic valve replacement approaches include open heart surgery, minimally invasive cardiac surgery (MICS) and minimally invasive catheter-based (percutaneous) aortic valve replacement. [29] [30] However, surgical aortic valve replacement is well-studied, and generally has a good and well-established longer-term prognosis. [31] A diseased aortic valve is most commonly replaced using a surgical procedure with either a mechanical or a tissue valve. ... "Effect of bisphosphonates on the progression of degenerative aortic stenosis". Echocardiography . 28 (1): 1–7. doi : 10.1111/j.1540-8175.2010.01256.x .
-
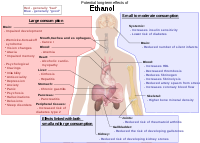
Long-Term Effects Of Alcohol
Wikipedia
In the UK a unit is defined as 10ml or 8g of pure alcohol. [28] Higher consumption increased overall mortality rate, but not above that of non-drinkers. [29] Other studies have found age-dependent mortality risks of low-to-moderate alcohol use: an increased risk for individuals aged 16–34 (due to increased risk of cancers, accidents, liver disease, and other factors), but a decreased risk for individuals ages 55+ (due to lower incidence of ischemic heart disease). [30] This is consistent with other research that found a J-curve dependency between alcohol consumption and total mortality among middle aged and older men.
-
Aneurysm
Wikipedia
A minority of aneurysms are caused by copper deficiency , which results in a decreased activity of the lysyl oxidase enzyme , affecting elastin , a key component in vessel walls. [25] [26] [27] Copper deficiency results in vessel wall thinning, [28] and thus has been noted as a cause of death in copper-deficient humans, [29] chickens and turkeys [30] Mechanics [ edit ] Aneurysmal blood vessels are prone to rupture under normal blood pressure and flow due to their special mechanical properties that make them weaker. ... "Renal artery an?eurysm: a report of 28 cases". Cardiovasc Surg . 4 (2): 185–189. doi : 10.1016/0967-2109(96)82312-X . ... International Journal for Numerical Methods in Biomedical Engineering . 28 (6–7): 801–808. doi : 10.1002/cnm.1481 .
-
Osteogenesis Imperfecta
Wikipedia
Thus far it seems to be limited to a First Nations people in Quebec . [25] Mutations in the gene CRTAP causes this type. [26] Type VIII [ edit ] OI caused by mutation in the gene LEPRE1 is classified as type VIII. [26] Type IX Osteogenesis imperfecta type IX (OI9) is caused by homozygous or compound heterozygous mutation in the PPIB gene on chromosome 15q22. [27] Type X Caused by homozygous mutation in the SERPINH gene on chromosome 11q13. [28] Type XI [ edit ] OI caused by mutations in FKBP10 on chromosome 17q21. [29] The mutations cause a decrease in secretion of trimeric procollagen molecules. ... Retrieved 2019-06-04 . ^ a b c d e f g Steiner RD, Pepin MG, Byers PH, Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP (January 28, 2005). " COL1A1/2 Osteogenesis Imperfecta" . ... "Phenotypic variability of osteogenesis imperfecta type V caused by an IFITM5 mutation" . J. Bone Miner. Res . 28 (7): 1523–30. doi : 10.1002/jbmr.1891 . ... Archived from the original on 2007-06-28 . Retrieved 2007-07-05 . ^ EL-Sobky, TA; Shawky, RM; Sakr, HM; Elsayed, SM; Elsayed, NS; Ragheb, SG; Gamal, R (15 November 2017). ... Archived from the original on 2007-09-28 . Retrieved 2007-07-05 . CS1 maint: bot: original URL status unknown ( link ) ^ "Osteogenesis Imperfecta Foundation | OIF.org" . www.oif.org .COL1A1, COL1A2, SERPINF1, CRTAP, P3H1, SPARC, WNT1, TMEM38B, PLOD2, P4HB, MESD, PPIB, SERPINH1, TENT5A, TAPT1, SMPD3, SMAD4, SUCO, CREB3L1, FKBP10, BMP1, IFITM5, SOST, PPP1R2C, BGLAP, DMD, BEST1, GH1, ACVR2B, SEC24D, PNPLA2, FGFR3, AGA2, PLS3, COX8A, DCN, LRP5, TLL1, PDIA2, GPATCH8, RER, WWTR1, MTCO2P12, PADI1, EFEMP2, PLA1A, MBTPS2, GPR180, NBAS, MED18, LINC01672, TRAP, MIR29B2, MIR29B1, MIR145, SP7, ACTB, EIF2AK3, TNC, ALPL, BAAT, BGN, TSPO, KRIT1, CD38, CD44, CHRM3, COL3A1, COL5A1, CSF2, DLX3, FN1, MSTN, IGF1, TNFSF11, LAMC2, LOX, LRP6, COX2, PPP1CB, PTH, PTGS2, RNASE1, SRSF2, SLC6A2, TNXB, TSC1, TSHR, CXCR4, LOC107984355
-
Spinal Cord Injury
Wikipedia
In younger people, it most commonly results from neck flexion. [26] The most common causes are falls and vehicle accidents; however other possible causes include spinal stenosis and impingement on the spinal cord by a tumor or vertebral disk. [27] Anterior cord syndrome [ edit ] Anterior cord syndrome , due to damage to the front portion of the spinal cord or reduction in the blood supply from the anterior spinal artery , can be caused by fractures or dislocations of vertebrae or herniated disks. [25] Below the level of injury, motor function, pain sensation, and temperature sensation are lost, while sense of touch and proprioception (sense of position in space) remain intact. [28] [26] These differences are due to the relative locations of the spinal tracts responsible for each type of function. [25] Brown-Séquard syndrome [ edit ] Brown-Séquard syndrome occurs when the spinal cord is injured on one side much more than the other. [29] It is rare for the spinal cord to be truly hemisected (severed on one side), but partial lesions due to penetrating wounds (such as gunshot or knife wounds) or fractured vertebrae or tumors are common. [30] On the ipsilateral side of the injury (same side), the body loses motor function, proprioception , and senses of vibration and touch. [29] On the contralateral (opposite side) of the injury, there is a loss of pain and temperature sensations. [27] [29] Posterior cord syndrome [ edit ] Posterior cord syndrome , in which just the dorsal columns of the spinal cord are affected, is usually seen in cases of chronic myelopathy but can also occur with infarction of the posterior spinal artery . [31] This rare syndrome causes the loss of proprioception and sense of vibration below the level of injury [26] while motor function and sensation of pain, temperature, and touch remain intact. [32] Usually posterior cord injuries result from insults like disease or vitamin deficiency rather than trauma. [33] Tabes dorsalis , due to injury to the posterior part of the spinal cord caused by syphilis, results in loss of touch and proprioceptive sensation. [34] Conus medullaris and cauda equina syndromes [ edit ] Conus medullaris syndrome is an injury to the end of the spinal cord, located at about the T12–L2 vertebrae in adults. [29] This region contains the S4–S5 spinal segments, responsible for bowel, bladder, and some sexual functions , so these can be disrupted in this type of injury. [29] In addition, sensation and the Achilles reflex can be disrupted. [29] Causes include tumors , physical trauma, and ischemia . [35] Cauda equina syndrome (CES) results from a lesion below the level at which the spinal cord splits into the cauda equina , [33] at levels L2–S5 below the conus medullaris. [36] Thus it is not a true spinal cord syndrome since it is nerve roots that are damaged and not the cord itself; however, it is common for several of these nerves to be damaged at the same time due to their proximity. [35] CES can occur by itself or alongside conus medullaris syndrome. [36] It can cause low back pain, weakness or paralysis in the lower limbs, loss of sensation, bowel and bladder dysfunction, and loss of reflexes. [36] Unlike in conus medullaris syndrome, symptoms often occur on only one side of the body. [35] The cause is often compression, e.g. by a ruptured intervertebral disk or tumor. [35] Since the nerves damaged in CES are actually peripheral nerves because they have already branched off from the spinal cord, the injury has better prognosis for recovery of function: the peripheral nervous system has a greater capacity for healing than the central nervous system . [36] Signs and symptoms [ edit ] Actions of the spinal nerves Level Motor Function C1 – C6 Neck flexors C1 – T1 Neck extensors C3 , C4 , C5 Supply diaphragm (mostly C4 ) C5 , C6 Move shoulder , raise arm ( deltoid ); flex elbow ( biceps ) C6 externally rotate ( supinate ) the arm C6 , C7 Extend elbow and wrist ( triceps and wrist extensors ); pronate wrist C7 , T1 Flex wrist; supply small muscles of the hand T1 – T6 Intercostals and trunk above the waist T7 – L1 Abdominal muscles L1 – L4 Flex thigh L2 , L3 , L4 Adduct thigh; Extend leg at the knee ( quadriceps femoris ) L4 , L5 , S1 abduct thigh; Flex leg at the knee ( hamstrings ); Dorsiflex foot ( tibialis anterior ); Extend toes L5 , S1 , S2 Extend leg at the hip ( gluteus maximus ); Plantar flex foot and flex toes Further information: Dermatome (anatomy) Signs (observed by a clinician) and symptoms (experienced by a patient) vary depending on where the spine is injured and the extent of the injury. ... Of the incomplete SCI syndromes, Brown-Séquard and central cord syndromes have the best prognosis for recovery and anterior cord syndrome has the worst. [28] People with nontraumatic causes of SCI have been found to be less likely to suffer complete injuries and some complications such as pressure sores and deep vein thrombosis, and to have shorter hospital stays. [11] Their scores on functional tests were better than those of people with traumatic SCI upon hospital admission, but when they were tested upon discharge, those with traumatic SCI had improved such that both groups' results were the same. [11] In addition to the completeness and level of the injury, age and concurrent health problems affect the extent to which a person with SCI will be able to live independently and to walk. [8] However, in general people with injuries to L3 or below will likely be able to walk functionally, T10 and below to walk around the house with bracing, and C7 and below to live independently. [8] New therapies are beginning to provide hope for better outcomes in patients with SCI, but most are in the experimental/translational stage. [3] One important predictor of motor recovery in an area is presence of sensation there, particularly pain perception. [36] Most motor recovery occurs in the first year post-injury, but modest improvements can continue for years; sensory recovery is more limited. [123] Recovery is typically quickest during the first six months. [124] Spinal shock , in which reflexes are suppressed, occurs immediately after the injury and resolves largely within three months but continues resolving gradually for another 15. [125] Sexual dysfunction after spinal injury is common.
-
Sex-Selective Abortion
Wikipedia
For example, Jiang et al. claim that the birth sex ratio in China was 116–121 over a 100-year period in the late 18th and early 19th centuries; in the 120–123 range in the early 20th century; falling to 112 in the 1930s. [24] [25] Data on human sex ratio at birth [ edit ] In the United States, the sex ratios at birth over the period 1970–2002 were 105 for the white non-Hispanic population, 104 for Mexican Americans, 103 for African Americans and Native Americans, and 107 for mothers of Chinese or Filipino ethnicity. [26] Among Western European countries c. 2001, the ratios ranged from 104 to 107. [27] [28] [29] In the aggregated results of 56 Demographic and Health Surveys [30] in African countries, the birth sex ratio was found to be 103, though there is also considerable country-to-country, and year-to-year variation. [31] In a 2005 study, U.S. ... According to a 2011 Gallup poll , if they were only allowed to have one child, 40% of respondents said they would prefer a boy, while only 28% preferred a girl. [114] When told about prenatal sex selection techniques such as sperm sorting and in vitro fertilization embryo selection, 40% of Americans surveyed thought that picking embryos by sex was an acceptable manifestation of reproductive rights. [115] These selecting techniques are available at about half of American fertility clinics, as of 2006. [116] However, other studies show a larger preference for females.
-
Food Allergy
Wikipedia
Balsam of Peru , which is in various foods, is in the "top five" allergens most commonly causing patch test reactions in people referred to dermatology clinics. [27] Sensitization [ edit ] Sensitization can occur through the gastrointestinal tract, respiratory tract and possibly the skin. [28] Damage to the skin in conditions such as eczema has been proposed as a risk factor for sensitization. [29] An Institute of Medicine report says that food proteins contained in vaccines, such as gelatin , milk, or egg can cause sensitization (development of allergy) in vaccine recipients, to those food items. [30] Atopy [ edit ] Food allergies develop more easily in people with the atopic syndrome , a very common combination of diseases: allergic rhinitis and conjunctivitis , eczema , and asthma . [31] The syndrome has a strong inherited component; a family history of allergic diseases can be indicative of the atopic syndrome. [ medical citation needed ] Cross-reactivity [ edit ] Some children who are allergic to cow's milk protein also show a cross-sensitivity to soy-based products. [32] Some infant formulas have their milk and soy proteins hydrolyzed, so when taken by infants, their immune systems do not recognize the allergen and they can safely consume the product. ... Asthma and Allergy Foundation of America. 28 March 2007. Archived from the original on 7 December 2006 . ... ISBN 978-0-8153-4101-7 . Archived from the original on 28 June 2009. ^ Grimbaldeston MA, Metz M, Yu M, et al. (2006).
-
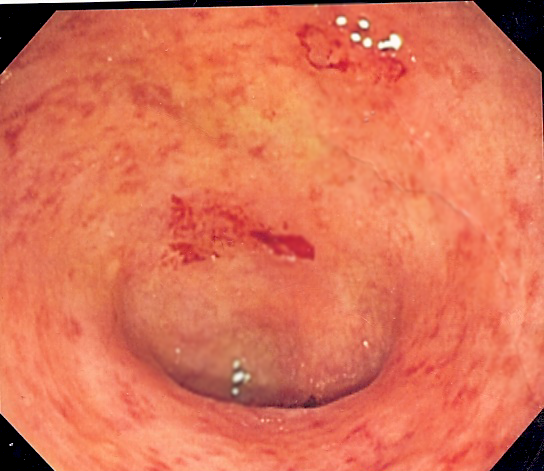
Ulcerative Colitis
Wikipedia
Commonly affected organ systems include: eyes, joints, skin, and liver. [26] The frequency of such extraintestinal manifestations has been reported as between 6 and 47%. [27] [28] UC may affect the mouth. About 8% of individuals with UC develop oral manifestations. [29] The two most common oral manifestations are aphthous stomatitis and angular cheilitis . [29] Aphthous stomatitis is characterized by ulcers in the mouth, which are benign, noncontagious and often recurrent.IL23R, NOD2, PTPN2, CCL20, IL1B, ICAM1, TNF, IRF5, HLA-DRB1, TNFSF15, FCGR2A, IL10, STAT3, MST1, IRGM, CARD9, IL7R, IL17REL, CCNY, ADCY7, GNA12, MMP9, GPR65, MASP2, DAP, ABCB1, CXCL8, PRDM1, CDH3, NKX2-3, IL12B, MPO, JAK2, CDH1, ECM1, RELA, HNF4A, CCR6, SLC11A1, ORMDL3, LAMB1, SLC26A3, HERC2, SPHK1, RIPK2, ARPC2, ICOSLG, IL1R2, IL10RA, CASP3, DEFA5, KIF21B, IL10RB, CXCR2, IL18RAP, PSMG1, GHRL, VCAM1, LTF, CXCR1, IKZF1, LSP1, PTGS2, DEFA6, MUC2, MMP1, FAS, PTGS1, COL18A1, TLR4, ADRB2, IL6, ATG16L1, FASLG, NFKB1, PTPN22, CD40, STAT4, IL27, NOS2, IL2RA, TNFRSF1A, SMAD3, CD28, SLC44A4, TCF4, HLA-DRA, TNFAIP3, FUT2, AHR, IL19, IL26, TNFRSF6B, IL6R, ETS1, UBE2L3, IL1R1, HLA-DQA1, CFB, ZNF365, GSDMB, ADCY3, ZFP90, TNFRSF14, ZMIZ1, BACH2, IFNGR2, IL18R1, IFNG-AS1, NFATC1, BSN, DNMT3A, KIAA1109, NOTCH1, ITGAL, PRRC2A, NR5A2, TYK2, CDKAL1, GPR35, FAM177A1, IL4, OTUD3, LURAP1L, SLC9A8, PROCR, RTF1, ATXN2L, ADGRL2, ANTXR2, CLEC16A, GCKR, GBAP1, IL9, IL1RN, TNIP1, CRB1, GLYAT, ACTR1A, TOM1, SH2B3, PNKD, IL13, IL17A, RMI2, KEAP1, NLRP3, IL17F, SETD1A, IL2, JAZF1, PARK7, ADAD1, CMC1, AMZ1, CIT, PUS10, HLA-DRB9, HORMAD2, SBNO2, FNBP1, FGFR1OP, PKIG, DENND1B, LINC01620, SLC39A11, HSPA1L, CD226, HLA-B, TBC1D9, RBM45, ZNF831, ACAD8, IFNG, ZPBP2, HIPK1, NRAD1, LRRK2, NXPE1, FNDC3A, IL1A, GPR18, RGS14, RAVER1, TNXB, NPEPPS, BRD2, RIC8B, INAVA, IGF2-AS, ATXN2, GATD3A, FOXP3, RPS6KB1, IBD5, PMP22, RDX, PTPRC, IL22, PTGIR, GABPA, PRKCQ, PRKCB, SFMBT1, SLC22A4, IL23A, BANK1, UBASH3A, ERAP1, RTEL1, TLR2, TGFB1, CCHCR1, TET2, SUOX, STK11, DPH5, HDAC7, INTS11, FAM118A, VDR, PPP2R3C, PPARG, PLCG2, ZNF300, CYTH1, CPEB4, UQCR10, NDFIP1, FIBP, DUSP16, OSMR, TSPAN14, ATG5, PLCL1, TP53, LITAF, ZGPAT, NKD1, TRIM15, IL33, ANKRD30A, SEMA6D, SP140, SKAP2, MFSD13A, PLAU, THADA, DELEC1, GAL3ST2, TMBIM1, IFIH1, KSR1, ERAP2, NFE2L2, ASAP2, NR1I2, COX2, MSH5, MPZ, ANKRD55, MAML2, CHP1, UBAC2, ALB, C5orf66, ARSA, MMP24OS, LINC01804, LINC01475, LINC01250, LINC00824, LINC01147, MTCO2P12, CELSR3, LINC00993, TEX41, LINC01185, TRAF3IP2-AS1, C1orf141, EEF1AKMT2, APEH, ERN1, NR0B1, LINC02863, CCR5AS, LINC00598, DAG1, LINC02213, DUPD1, CRP, LINC00484, CAMK2A, ACTA2, INS-IGF2, RTEL1-TNFRSF6B, SPATA48, CD6, C17orf67, IRF1-AS1, TMEM258, CTLA4, CDKN2A, CD14, MYRF, MSH5-SAPCD1, CYP21A2, CREM, MAPK1, BCL2, MLH1, IL21, MBL2, IL18, MICA, IL37, CCL11, DLG5, MIR21, MYO9B, MIR31, MYDGF, RUNX3, MIR155, JAK1, LCN2, IBD2, ESR1, TLR9, IL17D, MYD88, BPI, MIF, IBD3, GALM, GSTM1, VIP, CTNNB1, APC, STAT6, TPMT, IL5, GORASP1, TLR5, MSH2, S100A9, MTHFR, WNK1, SYT1, MIR146A, TSLP, CLDN1, IL7, CLDN2, CD80, ISG20, TFF3, KRAS, MADCAM1, CLDN7, TAC1, SOD1, XBP1, TRBV20OR9-2, VEGFA, MEFV, MMP7, CCR5, LTA, SPP1, NOD1, HMOX1, CDKN1A, AKT1, HPGDS, EGFR, EBI3, MGMT, NFKBIL1, CXCL12, PRTN3, TP63, SOCS1, LINC02605, PI3, ARMH1, CXCL5, CYP3A5, PLA2G1B, CCR9, CYP27B1, EBNA1BP2, RPSA, OCLN, HSP90B1, PSMD7, CCL2, H3P28, BTNL2, LEP, LGALS3, NFKBIA, GP2, REG4, LANCL1, CCN2, TRPV1, CA2, MMP10, FGF7, CABIN1, GSTP1, F5, NR3C1, MTFMT, SLC22A5, NAT2, DMBT1, RABEPK, GATA3, GSTT1, ARHGEF2, SOCS3, STAT1, MUC1, GSTK1, CCL26, BIN1, PAEP, F2RL1, CTF1, IRAK3, MAGI2, CXCR3, SLC25A20, KRT20, CD24, POLDIP2, SLC6A4, GDE1, CREBBP, TREM1, CRK, HLA-DRB3, TACR1, LACC1, TNFRSF1B, FHIT, CASP1, CAMP, MAPK14, PTPN11, CDKN2B-AS1, TIMP1, RNF19A, ACP1, TLR3, MIP, CLEC7A, CRHR1, MICB, HIF1A, HRAS, IL24, CDR3, S100B, LINC02210-CRHR1, MIR499A, AHSA1, TRAF3IP2, ST2, CXCL10, PPARA, PON1, DCC, ADORA3, PTGER4, PTGDS, IL15, CXCR6, ACE, ABCG2, DEFB4A, FCGR3A, IL1RL1, GRAP2, ATN1, SGSM3, IL4R, AQP8, CD163, S1PR1, SLCO6A1, PLA2G2A, CCL25, SERPINA1, LYZ, CX3CR1, MIR126, HGF, MAP2K7, SFTPD, DEFB4B, CXCR4, SELL, AIMP2, CD68, LAMC2, CD44, IBD6, HLA-DQB1, H3P10, SDC1, CX3CL1, NLRP12, MIR196A2, CEACAM5, H2AX, HAMP, ITGB1, RNF186, SOX2, SOD2, PAG1, S100A1, SFRP2, CCL5, CCL8, REG1A, CCL21, CCL24, REL, CYP26B1, CCL3, SFRP1, DEFB103B, SOAT1, OPN1LW, SLC9A3, SLC15A1, RAG2, SLC52A1, CD34, SLPI, CD86, S100A8, CDH11, MOK, STAT5A, SEMA3E, ALPI, SEC14L2, SMUG1, LY96, PANX1, ABCB6, APEX1, TAP2, NOX1, BIRC2, BIRC3, IL17B, CHST2, TCEAL1, SIRT1, IFITM3, AGER, NXF1, AGA, AFM, MPRIP, TNFSF13B, CORIN, ADORA2A, ADM, KLRK1, CARD8, RIPK3, CD160, RASSF1, SLC6A14, APOE, PER3, CFLAR, YWHAZ, TCF7L2, TCN2, TERT, CBR1, TGFBR2, EIM, TLR1, CBFA2T3, CAT, TNFAIP6, CASP9, TXN, CALCA, XRCC1, C4B, B2M, CRNKL1, RAC1, PLA2G6, KLRC4-KLRK1, USO1, RAB4B-EGLN2, CUZD1, MBTPS1, EED, RIPK1, IL20, CCND1, DUOX2, PSC, HSPA14, NAT1, MEPE, MIR215, LGALS4, MIR214, LEPR, CYP2C19, HAVCR2, MIR206, KIR3DL1, MIR19A, KDR, EGLN2, ITGAV, ITGAE, IRAK1, INSRR, IDO1, IL16, LRG1, LPL, BCL2L12, IL11, SMAD7, MUC5AC, MUC4, MUC3A, MTRR, CRH, COX1, MST1R, DEFB103A, MMP3, MMP2, MLN, MIR29A, MET, MEP1A, MDM2, MDK, ZBP1, TAGAP, DEFB1, MYOD1, HLA-G, PLB1, NPSR1, HLA-DPB1, HLA-C, HLA-A, LINC01193, GZMB, GUCA2B, GUCA2A, MLKL, FCGR3B, FGFR1, FOXO3, FLNB, FLT1, GEM, FOS, FAAH, NR4A1, NQO1, F2, MIR145, MIR143, EFNA5, MIR122, EIF4E, ELAVL2, ELF3, EPHB2, IDH1, TNC, HSPD1, HSP90AA1, HSPA4, DOCK11, MUC17, HT, CDAN1, MXI1, CYP2B6, MIR375, PCBP4, PSD, MUC3B, SLC11A2, CISH, COL2A1, MIR146B, MIR196B, CDX2, SLC12A9, NOS3, WG, TNFRSF11B, CHGA, PIK3CD, ODC1, PTPRS, PIK3CB, PIK3CA, CHI3L1, PIK3CG, PECAM1, PAK1, PKM, RNU1-1, TAAR1, B3GAT1, NSG1, SELENOS, SCYL1, SGPP2, CASZ1, LOC102723407, RIBC2, RPTOR, LANCL2, CHMP2B, MIR323B, LOC102724197, DEFB104A, TRIM58, NSFL1C, CBSL, AICDA, PART1, DNAJB5, H3P16, DUOXA2, BBC3, MIR137, MIR184, CLEC2D, ST6GALNAC1, DBNL, LAMTOR2, IGHD3-9, IGHD3-10, MIR150, MIR429, MIR448, MIR142, MIR141, IGHV3-69-1, IGHV3OR16-7, CPAMD8, KIAA1614, STK39, RABGEF1, MIR106A, MIR449A, PDCD4, EARS2, SNX20, LOC102724971, IL17C, MIB1, CGN, NAAA, LOC390714, CCNDBP1, IL34, BRINP3, TRPV3, ACKR3, LOC107987479, DNAJB1P1, PLBD2, MIR495, NLRP7, APOBEC3A, MUC20, ATF6, MAPRE1, USP12, LOC110806262, TCERG1L, MUC13, BAHD1, BMP6P1, SUCNR1, H19, CCDC88B, H3P13, H3P8, MGLL, OR10A4, STK38, LGALS14, ANKS1B, CHPT1, CST12P, SLC5A8, MIR490, KYAT3, YDJC, NUP62, AMACR, SLURP1, PADI4, COMMD1, LYRM4, ZNF281, TPTEP2-CSNK1E, CCL4L1, MACF1, RTL1, TIGAR, PARD3, GADL1, GPBAR1, ASPRV1, NCAPD3, PERCC1, FKBP15, NDUFS7, AKR1B10, LOC105379528, MARVELD2, PANX2, MLC1, CBLL2, ACSBG1, CD274, ZNF410, RMC1, HCG23, RBMXL1, CAMKMT, MIR34C, HDAC11, DHDDS, MIR34B, MIR551B, P2RY13, S1PR5, MARCHF1, TM6SF2, PROSER1, FAM217B, MIR26B, IBD22, SF3B6, SLC22A23, IBD24, LDAH, MPIG6B, MIR223, IBD26, MSMP, LINC00994, IGAN1, ANP32E, MED25, LARS1, NCKIPSD, MUL1, RNF128, EXOC3-AS1, MIR629, XRCC6P5, MUC5B, CALHM6, ABCG5, NXPE4, AFTPH, ARHGEF28, D2HGDH, HHIP, IL17RD, HPP1, UGT1A1, UGT1A7, MIR372, NUCKS1, IL25, DDIT4, PINK1, MIR650, RBM47, SMOX, TOLLIP, ZNF649, ATG9A, TERF2IP, MMP28, MKS1, FSD1, BRWD1, ARHGAP24, CHMP5, ISYNA1, MIR422A, MIR200C, MIR200B, IL17RB, ZNF77, GGTLC1, REEP6, CXCL16, EGLN3, CAVIN3, FERMT1, MIR195, MIR4728, AZIN2, MIR193A, CD207, NORAD, DEFB104B, RRM2B, HIF1A-AS2, APOBEC3A_B, CYGB, CD209, MARCHF3, FCRL3, SLC40A1, TBX21, BEST2, MDFIC, MIR192, IMPACT, F11R, CLEC4A, NT5C3A, ITCH, MAGEC2, FSD1L, TRPV2, POP5, GNB4, MAGT1, CHST15, TRPV4, H3P37, TLR8, RPAIN, TLR7, H3C9P, RAVER2, CLDN18, ACE2, TUBB6, LXN, CXADRP1, AADAT, DCTN4, ANGPTL4, IL17RC, DERL2, ASCC1, MFSD2A, ATAD1, CELIAC2, TGFB2, PADI2, MCL1, MSH6, GTF2H1, GUCY2C, GZMA, GZMM, H1-0, HCRT, HCRTR1, HDAC1, HIC1, HLA-DOA, HLA-DPA1, HLA-DQB2, HLA-DRB4, MR1, HMGB1, HNF4G, HNMT, HP, HPGD, AGFG1, HES1, HSD11B2, HSF2, HSPA1A, HSPA1B, HSPA2, CXCL2, CXCL1, GPX2, FUT4, F12, FABP2, PTK2B, FAP, FASN, FCAR, FCGR1A, FGF2, FHL2, FKBP5, FPR2, FUT3, GH1, GPX1, GHR, GHSR, GLB1, GLI1, GNAO1, GNG11, GPR12, GPR15, GPER1, GPR42, FFAR2, MKNK2, DNAJB1, HTR3A, HTR7, LGALS1, KIR2DL1, KIR2DL2, KIR2DS3, KIR2DS4, KLK1, KNG1, KRT1, KRT7, KRT8, LAIR1, STMN1, LCT, LGALS3BP, KCNN1, LGALS9, LRP6, LRP5, LSAMP, CYP4F3, LYN, MAD2L1, SMAD2, SMAD4, MAF, MAFD1, MAP6, KCNQ2, KCNMA1, IAPP, IL13RA2, ICAM2, IDUA, IFNA1, IFNA13, IFNGR1, IGF1, IGF1R, IGF2, IGHA1, IGHG3, IGLL2P, IL6ST, IL15RA, JUNB, TNFRSF9, ILK, ING1, INPP5D, INSR, IRAK2, IRF1, IRF4, ITGAM, ITGAX, ITPA, JAK3, F9, F3, ESR2, CALM1, BAX, BCKDHA, BCL3, TNFRSF17, BMP7, BRAF, BRS3, BUB1B, C4A, C4BPB, CA1, DDR1, CALM2, ATF4, CALM3, CAMK4, CASR, RUNX2, CBS, KYAT1, KRIT1, CCNE1, CD1A, CD19, MS4A1, CD33, ATOH1, ATF3, CD38, ALPP, AOC1, ACACA, ACAT2, ACHE, ADA, PLIN2, ADRA1A, ADRA2A, ADRA2B, AGT, AHSG, ALDOB, AMY1A, ASMT, AMY1B, AMY1C, ANXA1, ANXA2, APBA1, APOA4, APOH, AREG, ARG1, ARHGDIA, ARR3, ARRB2, CD37, CD40LG, ERBB2, DMRT1, CYP2A6, CYP2C9, CYP2C18, CYP2D6, CYP2E1, CYP3A4, CD55, DAO, DAPK1, DECR1, DLD, DMD, DPEP1, CXADR, DPP4, DSG2, DUSP1, GPR183, EDNRA, EGF, EGR1, ELANE, ENG, ENO1, EPHX1, STX2, CYBA, CTBS, CD47, CCR7, CD74, CDH13, CDKN2D, CFTR, CEACAM3, CEACAM7, CHEK1, CHRNA3, CLCN2, CLU, CCR1, CCR3, CMM, VCAN, CNR1, COL6A1, MAP3K8, COX8A, CLDN4, CPOX, CRHR2, CRX, CRYGD, CRYZ, CSF2, CSNK1E, MAZ, ME1, AKAP13, MGAT5, HAT1, GAS7, BECN1, MARCO, TNFSF10, TNFRSF11A, TNFRSF10C, IL1RL2, PROM1, APLN, PER2, ARHGEF7, VNN1, SQSTM1, TAX1BP1, TRPA1, PGLYRP1, MAP3K14, PSTPIP1, LATS1, NEURL1, COX7A2L, LPAR2, GPR55, UBE4A, PPIG, ADIPOQ, IFITM1, PIK3R3, DHX16, ZAP70, TUFM, TWIST1, UCP2, NR1H2, VCL, VIM, VIPR1, BEST1, VWF, WAS, WNT11, XRCC3, ZBTB16, NR0B2, FZD5, LAP, PLA2G7, FZD3, TFPI2, TUSC3, FOSL1, NCOA3, FZD1, FZD4, LOH19CR1, ULK1, COX5A, CYP7B1, NTN1, TXNIP, TRAIP, TLR6, KLF2, CITED2, TUBA1B, NDRG1, YAP1, PRMT5, CLEC10A, FBLN5, SLC35A1, CXCL13, KHDRBS1, CDK2AP2, PLK2, NEK6, CCL27, SUB1, PRDX3, MAPRE2, LILRB4, ADAMTS13, RPP14, BVES, FAM107A, WIF1, STUB1, POP7, LY86, P2RY14, ARHGEF6, PCYT1B, GAL3ST1, GDF15, TBPL1, CXCL14, CCL4L2, CLOCK, ISG15, DCAF1, BMS1, ELMO1, SLC23A1, GDF11, NR1I3, NR1H4, NR1D2, IL18BP, MUC12, ACTR3, NAMPT, PDZK1IP1, LPAR6, TNK2, TRIM13, DDX39A, TSC1, TRAF5, TRAF3, PIK3R1, P4HB, PRDX1, SERPINE1, PRKN, PCNA, PDCD1, PDE4A, SERPINF1, PF4, PGA5, PGD, SERPINE2, PLEK, OGG1, PNN, POMC, PREP, PRKAR1A, MAPK3, MAPK8, MAPK9, MAP2K2, HTRA1, PSG2, PSMA7, PTEN, ORM2, NTRK1, PTPN6, ND4, CD99, MME, MMP12, MMP13, MMP14, MNAT1, MPG, MSH3, MSI1, MT1A, CYTB, NUDT1, MTR, NTF3, MUC6, MUTYH, MYC, MYCN, CEACAM6, NEDD9, NELL1, NEU1, NFATC2, NGF, NPR1, NT5E, PTPN4, PTX3, TPM3, TERF2, SRC, SST, SSTR4, SULT1E1, AURKA, TACR2, ADAM17, TAP1, TBXAS1, HNF1B, TCF7, ZEB1, TFF1, SPG7, TFF2, TFRC, TG, TGFA, TGM2, TIA1, TIAL1, TIMP3, TJP1, TP73, TPD52L1, TPH1, SPN, SP1, RAD51, CLEC11A, RAP1A, RARRES2, PLAAT4, UPF1, RNASE2, RNASE3, RNU1-4, RPGR, RPLP2, RPS19, S100A12, SAT1, SCNN1B, SMPD1, SCNN1G, CCL4, CXCL6, SELE, SELPLG, SETMAR, SGK1, SLC1A5, SLC3A2, SLC9A1, SLC16A1, SMO, H3P40
-
Circulating Tumor Cell
Wikipedia
This low frequency, associated to difficulty of identifying cancerous cells, means that a key component of understanding CTCs biological properties require technologies and approaches capable of isolating 1 CTC per mL of blood, either by enrichment, or better yet with enrichment-free assays that identify all CTC subtypes in sufficiently high definition to satisfy diagnostic pathology image-quantity requirements in patients with a variety of cancer types. [18] To date CTCs have been detected in several epithelial cancers (breast, prostate, lung, and colon) [28] [29] [30] [31] and clinical evidences indicate that patients with metastatic lesions are more likely to have CTCs isolated.
-
Ebola
Wikipedia
Specialty Infectious disease Symptoms Fever , sore throat , muscular pain , headaches , diarrhoea , bleeding [1] Complications Low blood pressure from fluid loss [2] Usual onset Two days to three weeks post exposure [1] Causes Ebolaviruses spread by direct contact [1] Diagnostic method Finding the virus, viral RNA , or antibodies in blood [1] Differential diagnosis Malaria , cholera , typhoid fever , meningitis , other viral haemorrhagic fevers [1] Prevention Coordinated medical services, careful handling of bushmeat [1] Treatment Supportive care [1] Medication Atoltivimab/maftivimab/odesivimab (Inmazeb) Prognosis 25–90% mortality [1] Ebola , also known as Ebola virus disease ( EVD ) or Ebola hemorrhagic fever ( EHF ), is a viral hemorrhagic fever of humans and other primates caused by ebolaviruses . [1] Signs and symptoms typically start between two days and three weeks after contracting the virus with a fever , sore throat , muscular pain , and headaches . [1] Vomiting , diarrhoea and rash usually follow, along with decreased function of the liver and kidneys . [1] At this time, some people begin to bleed both internally and externally. [1] The disease has a high risk of death, killing 25% to 90% of those infected, with an average of about 50%. [1] This is often due to low blood pressure from fluid loss , and typically follows six to 16 days after symptoms appear. [2] The virus spreads through direct contact with body fluids , such as blood from infected humans or other animals. [1] Spread may also occur from contact with items recently contaminated with bodily fluids. [1] Spread of the disease through the air between primates , including humans, has not been documented in either laboratory or natural conditions. [3] Semen or breast milk of a person after recovery from EVD may carry the virus for several weeks to months. [1] [4] [5] Fruit bats are believed to be the normal carrier in nature , able to spread the virus without being affected by it. [1] Other diseases such as malaria , cholera , typhoid fever , meningitis and other viral haemorrhagic fevers may resemble EVD. [1] Blood samples are tested for viral RNA , viral antibodies or for the virus itself to confirm the diagnosis. [1] [6] Control of outbreaks requires coordinated medical services and community engagement. [1] This includes rapid detection, contact tracing of those who have been exposed, quick access to laboratory services, care for those infected, and proper disposal of the dead through cremation or burial. [1] [7] Samples of body fluids and tissues from people with the disease should be handled with special caution. [1] Prevention includes limiting the spread of disease from infected animals to humans by handling potentially infected bushmeat only while wearing protective clothing, and by thoroughly cooking bushmeat before eating it. [1] It also includes wearing proper protective clothing and washing hands when around a person with the disease. [1] An Ebola vaccine was approved in the United States in December 2019. [8] While there is no approved treatment for Ebola as of 2019 [update] , [9] two treatments ( REGN-EB3 and mAb114 ) are associated with improved outcomes. [10] Supportive efforts also improve outcomes. [1] This includes either oral rehydration therapy (drinking slightly sweetened and salty water) or giving intravenous fluids as well as treating symptoms. [1] Atoltivimab/maftivimab/odesivimab (Inmazeb) was approved for medical use in the United States in October 2020, for the treatment of infection caused by Zaire ebolavirus . [11] The disease was first identified in 1976, in two simultaneous outbreaks: one in Nzara (a town in South Sudan ) and the other in Yambuku ( Democratic Republic of the Congo ), a village relatively near the Ebola River from which the disease takes its name. [1] EVD outbreaks occur intermittently in tropical regions of sub-Saharan Africa . [1] From 1976 to 2012, the World Health Organization reports 24 outbreaks involving 2,387 cases with 1,590 deaths . [1] [12] The largest outbreak to date was the epidemic in West Africa , which occurred from December 2013 to January 2016, with 28,646 cases and 11,323 deaths. [13] [14] [15] It was declared no longer an emergency on 29 March 2016. [16] Other outbreaks in Africa began in the Democratic Republic of the Congo in May 2017, [17] [18] and 2018. [19] [20] In July 2019, the World Health Organization declared the Congo Ebola outbreak a world health emergency . [21] Contents 1 Signs and symptoms 1.1 Onset 1.2 Bleeding 1.3 Recovery and death 2 Cause 2.1 Virology 2.2 Transmission 2.3 Initial case 2.4 Reservoir 3 Pathophysiology 3.1 Immune system evasion 4 Diagnosis 4.1 Laboratory testing 4.2 Differential diagnosis 5 Prevention 5.1 Vaccines 5.2 Infection control 5.2.1 Caregivers 5.2.2 Patients and household members 5.2.3 Disinfection 5.2.4 General population 5.2.5 Bushmeat 5.2.6 Corpses, burial 5.2.7 Transport, travel, contact 5.2.8 Laboratory 5.3 Putting on protective equipment 5.4 Removing protective equipment 5.5 Isolation 5.6 Contact tracing 6 Management 6.1 Standard support 6.2 Intensive care 7 Prognosis 8 Epidemiology 8.1 1976 8.1.1 Sudan 8.1.2 Zaire 8.2 1995–2014 8.3 2013–2016 West Africa 8.3.1 2014 spread outside West Africa 8.4 2017 Democratic Republic of the Congo 8.5 2018 Équateur province 8.6 2018–2020 Kivu 8.7 2020 Équateur province 9 Society and culture 9.1 Weaponisation 9.2 Literature 10 Other animals 10.1 Wild animals 10.2 Domestic animals 10.3 Reston virus 11 Research 11.1 Treatments 11.2 Diagnostic tests 11.3 Disease models 12 See also 13 References 13.1 Bibliography 14 External links Signs and symptoms Signs and symptoms of Ebola [22] Onset The length of time between exposure to the virus and the development of symptoms ( incubation period ) is between two and 21 days, [1] [22] and usually between four and ten days. [23] However, recent estimates based on mathematical models predict that around 5% of cases may take longer than 21 days to develop. [24] Symptoms usually begin with a sudden influenza -like stage characterised by feeling tired , fever , weakness , decreased appetite , muscular pain , joint pain , headache, and sore throat. [1] [23] [25] [26] The fever is usually higher than 38.3 °C (101 °F). [27] This is often followed by nausea, vomiting, diarrhoea , abdominal pain, and sometimes hiccups . [26] [28] The combination of severe vomiting and diarrhoea often leads to severe dehydration . [29] Next, shortness of breath and chest pain may occur, along with swelling , headaches , and confusion . [26] In about half of the cases, the skin may develop a maculopapular rash , a flat red area covered with small bumps, five to seven days after symptoms begin. [23] [27] Bleeding In some cases, internal and external bleeding may occur. [1] This typically begins five to seven days after the first symptoms. [30] All infected people show some decreased blood clotting . [27] Bleeding from mucous membranes or from sites of needle punctures has been reported in 40–50% of cases. [31] This may cause vomiting blood , coughing up of blood , or blood in stool . [32] Bleeding into the skin may create petechiae , purpura , ecchymoses or haematomas (especially around needle injection sites). [33] Bleeding into the whites of the eyes may also occur. [34] Heavy bleeding is uncommon; if it occurs, it is usually in the gastrointestinal tract . [35] The incidence of bleeding into the gastrointestinal tract was reported to be ~58% in the 2001 outbreak in Gabon, [36] but in the 2014–15 outbreak in the US it was ~18%, [37] possibly due to improved prevention of disseminated intravascular coagulation . [29] Recovery and death Recovery may begin between seven and 14 days after first symptoms. [26] Death, if it occurs, follows typically six to sixteen days from first symptoms and is often due to low blood pressure from fluid loss . [2] In general, bleeding often indicates a worse outcome, and blood loss may result in death. [25] People are often in a coma near the end of life. [26] Those who survive often have ongoing muscular and joint pain, liver inflammation , and decreased hearing, and may have continued tiredness, continued weakness, decreased appetite, and difficulty returning to pre-illness weight. [26] [38] Problems with vision may develop. [39] It is recommended that survivors of EVD wear condoms for at least twelve months after initial infection or until the semen of a male survivor tests negative for Ebola virus on two separate occasions. [40] Survivors develop antibodies against Ebola that last at least 10 years, but it is unclear whether they are immune to additional infections. [41] Cause EVD in humans is caused by four of five viruses of the genus Ebolavirus . ... As of 8 May 2016 [update] , 28,646 suspected cases and 11,323 deaths were reported; [13] [186] however, the WHO said that these numbers may be underestimated. [187] Because they work closely with the body fluids of infected patients, healthcare workers were especially vulnerable to infection; in August 2014, the WHO reported that 10% of the dead were healthcare workers. [188] 2014 Ebola virus epidemic in West Africa In September 2014, it was estimated that the countries' capacity for treating Ebola patients was insufficient by the equivalent of 2,122 beds; by December there were a sufficient number of beds to treat and isolate all reported Ebola cases, although the uneven distribution of cases was causing serious shortfalls in some areas. [189] On 28 January 2015, the WHO reported that for the first time since the week ending 29 June 2014, there had been fewer than 100 new confirmed cases reported in a week in the three most-affected countries. ... His condition worsened and he returned to the hospital on 28 September, where he died on 8 October.SARS1, ERVK-6, SARS2, ERVK-20, NPC1, ERVW-1, TLR4, TIMELESS, RBBP6, ARHGEF5, HAVCR1, ERVK-32, STAT1, PLAAT4, IFNG, TNF, NEDD4, BST2, GP2, ZNF415, IL10, BAG3, IL6, IFNA13, LSAMP, ATM, ROBO3, ANXA5, IFNB1, IFNA1, PIKFYVE, DDX58, LAMP3, PRKRA, CD209, TPCN2, SLC25A19, IFI30, RUVBL2, LGR6, HAMP, APEX2, MAVS, PELI1, ZC3HAV1, TLR7, CLEC10A, VAC14, MERTK, HSPA14, CASZ1, CPVL, NUSAP1, LPAR3, RETREG1, TLR9, SEPTIN8, MUL1, SMYD3, MIR155, ANO6, GPRC6A, MRGPRX1, CLEC4G, CCL4L1, VHLL, MIR320A, CBLL2, VN1R17P, GPR166P, MIR196B, MIR1246, ECT, LINC01672, OXER1, GPR151, GNPTAB, HAVCR2, IRF9, FUZ, LNPK, HM13, RILP, NTPCR, DISP1, TRIM6, TIMD4, PRDM6, NLRP3, MRGPRX3, MRGPRX4, RLN3, IFITM3, ACP2, ISG15, IL18, HLA-B, HMOX1, HSPA5, HSPD1, IL1B, IL4, IRF1, GP5, IRF7, ITPA, JUN, JUNB, JUND, KPNA1, HLA-A, GALNT1, GTPBP1, EGI, AGRP, ALB, CDSN, CSF2, DAG1, DPAGT1, EIF5A, FOSB, ESR1, FCGR1A, FCGR1B, FOLR1, FOLR2, FOS, KPNA5, LAMP1, LGALS1, ACTB, VTN, XRCC5, USP7, CCDC6, FZD4, RUVBL1, MBTPS1, NHS, RAB11A, ARTN, MYOM2, CD163, SH3BP5, CCL4L2, VEGFA, TYROBP, TSG101, TPI1, TOP1, TMPRSS2, TLR3, THBS1, STAU1, SIGLEC1, SFTPD, SDC1, CCL5, CCL4, PTPA, POMC, PRKN, SOCS1
-
Bovine Spongiform Encephalopathy
Wikipedia
After further development and testing, this method could be of great value in surveillance as a blood- or urine-based screening test for BSE. [25] [26] Classification [ edit ] BSE is a transmissible disease that primarily affects the central nervous system ; it is a form of transmissible spongiform encephalopathy , like Creutzfeldt–Jakob disease and kuru in humans and scrapie in sheep, and chronic wasting disease in deer . [17] [27] [28] Prevention [ edit ] A ban on feeding meat and bone meal to cattle has resulted in a strong reduction in cases in countries where the disease has been present. ... S2CID 1185101 . ^ "Creekstone Farms response to USDA appeal of summary judgement" (Press release). 3buddies. 30 May 2007. Archived from the original on 28 September 2007 . Retrieved 20 June 2009 . ^ Seltzer M (12 July 2008). ... Retrieved 18 October 2018 . ^ a b "Europe's mad cows" . The Economist . 28 November 2000 . Retrieved 23 September 2017 . ^ a b "End to 10-year British beef ban" . ... Clinical and Investigative Medicine . 28 (3): 101–4. PMID 16021982 . ^ Digesta Artis Mulomedicinae , Publius Flavius Vegetius Renatus External links [ edit ] Bovine spongiform encephalopathy at Wikipedia's sister projects Definitions from Wiktionary Media from Wikimedia Commons News from Wikinews Data from Wikidata Taxonomy from Wikispecies Bovine spongiform encephalopathy at Curlie OIE – World Organisation for Animal Health: BSE situation in the world and annual incidence rate UK BSE Inquiry Website, Archived at The National Archives v t e Beef and veal Production Argentine beef Beef cattle Cow–calf operation Feeder cattle Kobe beef Organic beef Products Cuts Baseball steak Blade steak Brisket Chateaubriand steak Chuck steak Fajita Filet mignon Flank steak Flap steak Hanger steak Plate Ranch steak Restructured steak Rib eye Rib steak Round Rump Short loin Short ribs Shoulder tender Sirloin Top sirloin Skirt steak Standing rib roast Strip Shank T-bone Tenderloin Tri-tip Trotters Tail Processed Jerky Aged Bresaola Cabeza Corned beef Frankfurter Rindswurst Ground Montreal smoked Pastrami Meat extract Offal Brain Heart Tongue Tendon Tripas Tripe Testicles Dishes Beefsteak List of steak dishes Balbacua Blanquette de veau Beef Wellington Beef bourguignon Beef bun Beef Manhattan Beef noodle soup Beef on weck Beef Stroganoff Boiled beef Bulgogi Bulalo Cachopo Calf's liver and bacon Cansi Chateaubriand (dish) Cheesesteak Chicken fried steak Cordon bleu Dendeng Feu French dip Ginger beef Galbi Gored gored Gyūdon Hamburg steak Hortobágyi palacsinta Iga penyet Italian beef Jellied veal Kalio Karađorđeva šnicla Kitfo London broil Mongolian beef Neobiani Ossobuco Pares Pho Pot roast Pozharsky cutlet P'tcha Ragout fin Rawon Rendang Roast beef Roast beef sandwich Salisbury steak Saltimbocca Selat solo Sha cha beef Suea rong hai Shooter's sandwich Steak and kidney pudding Steak Diane Steak and eggs Steak and oyster pie Steak au poivre Tartare Tafelspitz Tongseng Veal Milanese Veal Orloff Veal Oscar Vitello tonnato Wallenbergare Related meats American bison Beefalo Water buffalo Żubroń Other Bovine spongiform encephalopathy Beef hormone controversy Beef ring Carcass grade Darkcutter Meat on the bone Ractopamine - Beef USA beef imports Japan Taiwan South Korea ( 2008 US beef protest in South Korea ) v t e Prion diseases and transmissible spongiform encephalopathy Prion diseases in humans inherited/ PRNP : fCJD Gerstmann–Sträussler–Scheinker syndrome Fatal familial insomnia sporadic: sCJD Sporadic fatal insomnia Variably protease-sensitive prionopathy acquired/ transmissible: iCJD vCJD Kuru Prion diseases in other animals Bovine spongiform encephalopathy Camel spongiform encephalopathy Scrapie Chronic wasting disease Transmissible mink encephalopathy Feline spongiform encephalopathy Exotic ungulate encephalopathy v t e Eradication of infectious diseases Eradication of human diseases Successful Smallpox / Alastrim ( Eradication of smallpox ) Underway (global) Dracunculiasis ( Eradication of dracunculiasis ) Poliomyelitis ( Eradication of poliomyelitis ) Malaria ( Eradication of malaria ) Yaws ( Eradication of yaws ) Underway (regional) Hookworm Lymphatic filariasis Measles vaccine epidemiology Rubella Trachoma Onchocerciasis Syphilis Rabies Eradication of agricultural diseases Successful Rinderpest ( Eradication of rinderpest ) Underway Ovine rinderpest Bovine spongiform encephalopathy Eradication programs Global Global Polio Eradication Initiative Global Certification Commission Malaria Eradication Scientific Alliance Regional United States Boll Weevil Eradication Program National Malaria Eradication Program India India National PolioPlus Pulse Polio Poliomyelitis in Pakistan Every Last Child The Final Inch Related topics Globalization and disease Mathematical modelling of disease Pandemic Transmission horizontal vertical Vaccination Zoonosis Authority control BNF : cb123825025 (data) NDL : 00907989